Science Forum: The Brazilian Reproducibility Initiative
Abstract
Most efforts to estimate the reproducibility of published findings have focused on specific areas of research, even though science is usually assessed and funded on a regional or national basis. Here we describe a project to assess the reproducibility of findings in biomedical science published by researchers based in Brazil. The Brazilian Reproducibility Initiative is a systematic, multicenter effort to repeat between 60 and 100 experiments: the project will focus on a set of common methods, repeating each experiment in three different laboratories from a countrywide network. The results, due in 2021, will allow us to estimate the level of reproducibility of biomedical science in Brazil, and to investigate what aspects of the published literature might help to predict whether a finding is reproducible.
https://doi.org/10.7554/eLife.41602.001Introduction
Concerns about the reproducibility of published results in certain areas of biomedical research were initially raised by theoretical models (Ioannidis, 2005a), systematic reviews of the existing literature (Ioannidis, 2005b) and alarm calls by the pharmaceutical industry (Begley and Ellis, 2012; Prinz et al., 2011). These concerns have subsequently been covered both in scientific journals (Baker, 2016) and in the wider media (Economist, 2013; Harris, 2017). While funding agencies have expressed concerns about reproducibility (Collins and Tabak, 2014), efforts to replicate published findings in specific areas of research have mostly been conducted by bottom-up collaborations and supported by private funders. The Reproducibility Project: Psychology, which systematically reproduced 100 articles in psychology (Open Science Collaboration, 2015), was followed by similar initiatives in the fields of experimental economics (Camerer et al., 2016), philosophy (Cova et al., 2018) and social sciences (Camerer et al., 2018), with replication rates ranging between 36 and 78%. Two projects in cancer biology (both involving the Center for Open Science and Science Exchange) are currently ongoing (Errington et al., 2014; Tan et al., 2015).
Although such projects are very welcome, they are all limited to specific research topics or communities. Moreover, apart from the projects in cancer biology, most have focused on areas of research in which experiments are relatively inexpensive and straightforward to perform: this means that the reproducibility of many areas of biomedical research has not been studied. Furthermore, although scientific research is mostly funded and evaluated at a regional or national level, the reproducibility of research has not, to our knowledge, been studied at these levels. To begin to address this gap, we have obtained funding from the Serrapilheira Institute, a recently created nonprofit institution, in order to systematically assess the reproducibility of biomedical research in Brazil.
Our aim is to replicate between 60 and 100 experiments from life sciences articles published by researchers based in Brazil, focusing on common methods and performing each experiment at multiple sites within a network of collaborating laboratories in the country. This will allow us to estimate the level of reproducibility of research published by biomedical scientists in Brazil, and to investigate if there are aspects of the published literature that can help to predict whether a finding is reproducible.
Brazilian science in a nutshell
Scientific research in Brazil started to take an institutional form in the second half of the 20th century, despite the earlier existence of important organizations such as the Brazilian Academy of Sciences (established in 1916) and the Universities of Brazil (later the Federal University of Rio de Janeiro) (1920) and São Paulo (1934). In 1951, the federal government created the first national agency dedicated to funding research (CNPq), as well as a separate agency to oversee postgraduate studies (CAPES), although graduate-level education was not formalized in Brazil until 1965 (Schwartzman, 2001). CNPq and CAPES remain the major funders of Brazilian academic science.
As the number of researchers increased, CAPES took up on the challenge of creating a national evaluation system for graduate education programs in Brazil (Barata, 2016). In the 1990s, the criteria for evaluation began to include quantitative indicators, such as numbers of articles published. In 1998, significant changes were made with the aim of trying to establish articles in international peer-reviewed journals as the main goal, and individual research areas were left free to design their own criteria for ranking journals. In 2007, amidst the largest-ever expansion in the number of federal universities, the journal ranking system in the life sciences became based on impact factors for the previous year, and remains so to this day (CAPES, 2016).
Today, Brazil has over 200,000 PhDs, with more than 10,000 graduating every year (CGEE, 2016). Although the evaluation system is seen as an achievement, it is subject to much criticism, revolving around the centralizing power of CAPES (Hostins, 2006) and the excessive focus on quantitative metrics (Pinto and Andrade, 1999). Many analysts criticize the country’s research as largely composed of "salami science", growing in absolute numbers but lacking in impact, originality and influence (Righetti, 2013). Interestingly, research reproducibility has been a secondary concern in these criticisms, and awareness of the issue has begun to rise only recently.
With the economic and political crisis afflicting the country since 2014, science funding has suffered a sequence of severe cuts. As the Ministry for Science and Technology was merged with that of Communications, a recent constitutional amendment essentially froze science funding at 2016 levels for 20 years (Angelo, 2016). The federal budget for the Ministry suffered a 44% cut in 2017 and reached levels corresponding to roughly a third of those invested a decade earlier (Floresti, 2017), leading scientific societies to position themselves in defense of research funding (SBPC, 2018). Concurrently, CAPES has initiated discussions on how to reform its evaluation system (ABC, 2018). At this delicate moment, in which a new federal government has just taken office, an empirical assessment of the country’s scientific output seems warranted to inform such debates.
The Brazilian Reproducibility Initiative: aims and scope
The Brazilian Reproducibility Initiative was started in early 2018 as a systematic effort to evaluate the reproducibility of Brazilian biomedical science. Openly inspired by multicenter efforts such as the Reproducibility Project: Psychology (Open Science Collaboration, 2015), the Reproducibility Project: Cancer Biology (Errington et al., 2014) and the Many Labs projects (Ebersole et al., 2016; Klein et al., 2014; Klein et al., 2018), our goal is to replicate between 60 and 100 experiments from published Brazilian articles in the life sciences, focusing on common methods and performing each experiment in multiple sites within a network of collaborating laboratories. The project’s coordinating team at the Federal University of Rio de Janeiro is responsible for the selection of methods and experiments, as well as for the recruitment and management of collaborating labs. Experiments are set to begin in mid-2019, in order for the project to achieve its final results by 2021.
Any project with the ambition of estimating the reproducibility of a country’s science is inevitably limited in scope by the expertise of the participating teams. We will aim for the most representative sample that can be achieved without compromising feasibility, through the use of the strategies described below. Nevertheless, representativeness will be limited by the selected techniques and biological models, as well as by our inclusion and exclusion criteria – which include the cost and commercial availability of materials and the expertise of the replicating labs.
Focus on individual experiments
Our first choice was to base our sample on experiments rather than articles. As studies in basic biomedical science usually involve many experiments with different methods revolving around a hypothesis, trying to reproduce a whole study, or even its main findings, can be cumbersome for a large-scale initiative. Partly because of this, the Reproducibility Project: Cancer Biology (RP:CB), which had originally planned to reproduce selected main findings from 50 studies, has been downsized to fewer than 20 (Kaiser, 2018). Moreover, in some cases RP:CB has been able to reproduce parts of a study but has also obtained results that cannot be interpreted or are not consistent with the original findings. Furthermore, the individual Replication Studies published by RP:CB do not say if a given replication attempt has been successful or not: rather, the project uses multiple measures to assess reproducibility.
Contrary to studies, experiments have well defined effect sizes, and although different criteria can be used for what constitutes a successful replication (Goodman et al., 2016; Open Science Collaboration, 2015), they can be defined objectively, allowing a quantitative assessment of reproducibility. Naturally, there is a downside in that replication of a single experiment is usually not enough to confirm or refute the conclusions of an article (Camerer et al., 2018). However, if one’s focus is not on the studies themselves, but rather on evaluating reproducibility on a larger scale, we believe that experiments represent a more manageable unit than articles.
Selection of methods
No replication initiative, no matter how large, can aim to reproduce every kind of experiment. Thus, our next choice was to limit our scope to common methodologies that are widely available in the country, in order to ensure that we will have a large enough network of potential collaborators. To provide a list of candidate methods, we started by performing an initial review of a sample of articles in Web of Science life sciences journals published in 2017, filtering for papers which: a) had all authors affiliated with a Brazilian institution; b) presented experimental results on a biological model; c) did not use clinical or ecological samples. One hundred randomly selected articles had data extracted concerning the models, experimental interventions and methods used to analyze outcomes: the main results are shown in Figure 1A and B. A more detailed protocol for this step is available at https://osf.io/f2a6y/.
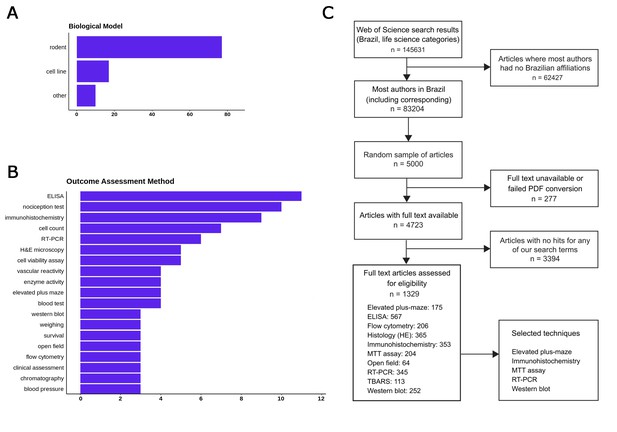
Selecting methods and papers for replication in the Brazilian Reproducibility Initiative.
(A) Most frequent biological models used in main experiments within a sample of 100 Brazilian life sciences articles. (B) Most frequent methods used for quantitative outcome detection in these experiments. ‘Cell count’, ‘enzyme activity’ and ‘blood tests’ include various experiments for which methodologies vary and/or are not described fully in articles. Nociception tests, although frequent, were not considered for replication due to animal welfare considerations. (C) Flowchart describing the first full-text screening round to identify articles in our candidate techniques, which led us to select our final set of five methods.
Based on this initial review, we restricted our scope to experiments using rodents and cell lines, which were by far the most prevalent models (present in 77 and 16% of articles, respectively). After a first round of automated full-text assessment of 5000 Brazilian articles between 1998 and 2017, we selected 10 commonly used techniques (Figure 1C) as candidates for replication experiments. An open call for collaborating labs within the country was then set up, and labs were allowed to register through an online form for performing experiments with one or more of these techniques and models during a three-month period. After this period, we used this input (as well as other criteria such as cost analysis) to select five methods for the replication effort: MTT assay, reverse transcriptase polymerase chain reaction (RT-PCR), elevated plus maze, western blot and immunohisto/cytochemistry (see https://osf.io/qxdjt/ for details). We are starting the project with the first three methods, while inclusion of the latter two will be confirmed after a more detailed cost analysis based on the fully developed protocols.
We are currently selecting articles using these techniques by full-text screening of a random sample of life sciences articles from the past 20 years in which most of the authors, including the corresponding one, are based in a Brazilian institution. From each of these articles, we select the first experiment using the technique of interest, defined as a quantitative comparison of a single outcome between two experimental groups. Although the final outcome of the experiment should be assessed using the method of interest, other laboratory techniques are likely to be involved in the model and experimental procedures that precede this step.
We will restrict our sample to experiments that: a) represent one of the main findings of the article, defined by mention of its results in the abstract; b) present significant differences between groups, in order to allow us to perform sample size calculations; c) use commercially available materials; d) have all experimental procedures falling within the expertise of at least three laboratories in our network; e) have an estimated cost below 0.5% of the project’s total budget. For each included technique, 20 experiments will be selected, with the biological model and other features of the experiment left open to variation in order to maximize representativeness. A more detailed protocol for this step is available at https://osf.io/u5zdq/.
After experiments are selected, we will record each study’s methods description in standardized description forms, which will be used to define replication protocols. These experiments will then be assigned to three laboratories each by the coordinating team, which will confirm that they have the necessary expertise in order to perform it.
Multicenter replication
A central tenet of our project is that replication should be performed in multiple laboratories. As discussed in other replication projects (Errington et al., 2014; Gilbert et al., 2016; Open Science Collaboration, 2015) a single failed replication is not enough to refute the original finding, as there are many reasons that can explain discrepancies between results (Goodman et al., 2016). While some of them – such as misconduct or bias in performing or analyzing the original experiment – are problematic, others – such as unrecognized methodological differences or chance – are not necessarily as alarming. Reproducibility estimates based on single replications cannot distinguish between these causes, and can thus be misleading in terms of their diagnoses (Jamieson, 2018).
This problem is made worse by the fact that data on inter-laboratory variability for most methods is scarce: even though simulations demonstrate that multicenter replications are an efficient way to improve reproducibility (Voelkl et al., 2018), they are exceedingly rare in most fields of basic biomedical science. Isolated attempts at investigating this issue in specific fields have shown that, even when different labs try to follow the same protocol, unrecognized methodological variables can still lead to a large amount of variation (Crabbe et al., 1999; Hines et al., 2014; Massonnet et al., 2010). Thus, it might be unrealistic to expect that reproducing a published experiment – for which protocol details will probably be lacking (Hair et al., 2018; Kilkenny et al., 2009) – will yield similar results in a different laboratory.
In our view, the best way to differentiate irreproducibility due to bias or error from that induced by methodological variables alone is to perform replications at multiple sites. In this way, an estimate of inter-laboratory variation can be obtained for every experiment, allowing one to analyze whether the original result falls within the expected variation range. Multicenter approaches have been used successfully in the area of psychology (Ebersole et al., 2016; Klein et al., 2014; Klein et al., 2018), showing that some results are robust across populations, while others do not reproduce well in any of the replication sites.
Our plan for the Brazilian Reproducibility Initiative is to perform each individual replication in at least three different laboratories; this, however, opens up questions about how much standardization is desirable. Although one should follow the original protocol in a direct replication, there are myriad steps that will not be well described. And while some might seem like glaring omissions, such as the absence of species, sex and age information in animal studies (Kilkenny et al., 2009), others might simply be overlooked variables: for example, how often does one describe the exact duration and intensity of sample agitation (Hines et al., 2014)? When conditions are not specified, one is left with two choices. One of them is to standardize steps as much as possible, building a single, detailed replication protocol for all labs. However, this will reduce inter-laboratory variation to an artificially low level, making the original experiment likely to fall outside the effect range observed in the replications.
To avoid this, we will take a more naturalistic approach. Although details included in the original article will be followed explicitly in order for the replication to be as direct as possible, steps which are not described will be left open for each replication team to fill based on their best judgment. Replication teams will be required to record those choices in detailed methods description forms, but it is possible – and desirable – for them to vary according to each laboratory’s experience. Methodological discrepancies in this case should approach those observed between research groups working independently, providing a realistic estimate of inter-laboratory variation for the assessment of published findings. This approach will also allow us to explore the impact of methodological variation on the experimental results – a topic perhaps as important as reproducibility itself – as a secondary outcome of the project.
Protocol review
A central issue in other replication projects has been engagement with the original authors in order to revise protocols. While we feel this is a worthy endeavor, the rate of response to calls for sharing protocols, data or code is erratic (Hardwicke and Ioannidis, 2018; Stodden et al., 2018; Wicherts et al., 2011). Moreover, having access to unreported information is likely to overestimate the reproducibility of a finding based on published information, leading results to deviate from a ‘naturalistic’ estimate of reproducibility (Coyne, 2016). Thus, although we will contact the original authors for protocol details when these are available, in order to assess methodological variation between published studies and replications, this information will not be made available to the replication teams. They will receive only the protocol description from the published article, with no mention of its results or origin, in order to minimize bias. While we cannot be sure that this form of blinding will be effective, as experiments could be recognizable by scientists working in the same field, replicating labs will be stimulated not to seek this information.
Lastly, although non-described protocol steps will be left open to variation, methodological issues that are consensually recognized to reduce error and bias will be enforced. Thus, bias control measures such as blinding of researchers to experimental groups will be used whenever possible, and sample sizes will be calculated to provide each experiment with a power of 95% to detect the original difference – as in other surveys, we are setting our power estimates at a greater than usual rate due to the recognition that the original results are likely to be inflated by publication bias. Moreover, if additional positive and/or negative controls are judged to be necessary to interpret outcomes, they will also be added to the experiment.
To ensure that these steps are followed – as well as to adjudicate on any necessary protocol adaptations, such as substitutions in equipment or materials – each individual protocol will be reviewed after completion in a round-robin approach (Silberzahn et al., 2018) by (i) the project’s coordinating team and (ii) an independent laboratory working with the same technique that is not directly involved in the replication. Each of the three protocol versions of every experiment will be sent to a different reviewing lab, in order to minimize the risk of over-standardization. Suggestions and criticisms to the protocol will be sent back to the replicating team, and experiments will only start after both labs and the coordinating team reach consensus that the protocol: a) does not deviate excessively from the published one and can be considered a direct replication; b) includes measures to reduce bias and necessary controls to ensure the validity of results.
Evaluating replications
As previous projects have shown, there are many ways to define a successful replication, all of which have caveats. Reproducibility of the general conclusions on the existence of an effect (e.g. two results finding a statistically significant difference in the same direction) might not be accompanied by reproducibility of the effect size; conversely, studies with effect sizes that are similar to each other might have different outcomes in significance tests (Simonsohn, 2015). Moreover, if non-replication occurs, it is hard to judge whether the original study or the replication is closer to the true result. Although one can argue that, if replications are conducted in an unbiased manner and have higher statistical power, they are more likely to be accurate, the possibility of undetected methodological differences preclude one from attributing non-replication to failures in the original studies.
Multisite replication is a useful way to circumvent some of these controversies, as if the variation between unbiased replications in different labs is known, it is possible to determine whether the original result is within this variability range. Thus, the primary outcome of our analysis will be the percentage of original studies with effect sizes falling within the 95% prediction interval of a meta-analysis of the three replications. Nevertheless, we acknowledge that this definition also has caveats: if inter-laboratory variability is high, prediction intervals can be wide, leading a large amount of results to be considered “reproducible”. Thus, replication estimates obtained by these methods are likely to be optimistic. On the other hand, failed replications will be more likely to reflect true biases, errors or deficiencies in the original experiments (Patil et al., 2016).
An additional problem is that, given our naturalistic approach to reproducibility, incomplete reporting in the original study might increase inter-laboratory variation and artificially improve our primary outcome. With this in mind, we will include other ways to define reproducibility as secondary outcomes, such as the statistical significance of the pooled replication studies, the significance of the effect in a meta-analysis including the original result and replication attempts, and a statistical comparison between the pooled effect sizes of the replications and the original result. We will also examine thoroughness of methodological reporting as an independent outcome, in order to evaluate the possibility of bias caused by incomplete reporting.
Moreover, we will explore correlations between results and differences in particular steps of each technique; nevertheless, we cannot know in advance whether methodological variability will be sufficient to draw conclusions on these issues. As each experiment will be performed in only three labs, while there are myriad steps to each technique, it is unlikely that we will be able to pinpoint specific sources of variation between results of individual experiments. Nevertheless, by quantifying the variation across protocols for the whole experiment, as well as for large sections of it (model, experimental intervention, outcome detection), we can try to observe whether the degree of variation at each level correlates with variability in results. Such analyses, however, will only be planned once protocols are completed, so as to have a better idea of the range of variability across them.
Finally, we will try to identify factors in the original studies that can predict reproducibility, as such proxies could be highly useful to guide the evaluation of published science. These will include features shown to predict reproducibility in previous work, such as effect sizes, significance levels and subjective assessment by prediction markets (Dreber et al., 2015; Camerer et al., 2016; Camerer et al., 2018; Open Science Collaboration, 2015); the pool of researchers used for the latter, however, will be different from those performing replications, so as not to compromise blinding with respect to study source and results. Other factors to be investigated include: a) the presence of bias control measures in the original study, such as blinding and sample size calculations; b) the number of citations and impact factor of the journal; c) the experience of the study’s principal investigator; d) the Brazilian region of origin; e) the technique used; f) the type of biological model; g) the area of research. As our sample of experiments will be obtained randomly, we cannot ensure that there will be enough variability in all factors to explore them meaningfully. Nevertheless, we should be able to analyze some variables that have not been well explored in previous replication attempts, such as ‘impact’ defined by citations and publication venues, as most previous studies have focused on particular subsets of journals (Camerer et al., 2018; Open Science Collaboration, 2015) or impact tiers (Errington et al., 2014; Ioannidis, 2005b).
A question that cannot be answered directly by our study design is whether any correlations found in our sample of articles can be extrapolated either to different methods in Brazilian biomedical science or to other regions of the world. For some factors, including the reproducibility estimates themselves and their correlation with local variables, extrapolations to the international scenario are clearly not warranted. On the other hand, relationships between reproducibility and methodological variables, as well as with article features, can plausibly apply to other countries, although this can only be known for sure by performing studies in other regions.
All of our analyses will be preregistered at the Open Science Framework in advance of data collection. All our datasets will be made public and updated progressively as replications are performed – a process planned to go on until 2021. As an additional measure to promote transparency and engage the Brazilian scientific community in the project, we are posting our methods description forms for public consultation and review (see http://reprodutibilidade.bio.br/public-consultation), and will do so for the analysis plan as well.
Potential challenges
A multicenter project involving the replication of experiments in multiple laboratories across a country of continental proportions is bound to meet challenges. The first of them is that the project is fully dependent on the interest of Brazilian laboratories to participate. Nevertheless, the response to our first call for collaborators exceeded our expectations, reaching a total of 71 laboratories in 43 institutions across 19 Brazilian states. The project received coverage by the Brazilian media (Ciscati, 2018; Neves and Amaral, 2018; Pesquisa FAPESP, 2018) and achieved good visibility in social networks, contributing to this widespread response. While we cannot be sure that all laboratories will remain in the project until its conclusion, it seems very likely that we will have the means to perform our full set of replications, particularly as laboratories will be funded for their participation.
Concerns also arise from the perception that replicating other scientists’ work indicates mistrust of the original results, a problem that is potentiated by the conflation of the reproducibility debate with that on research misconduct (Jamieson, 2018). Thus, from the start, we are taking steps to ensure that the project is viewed as we conceive it: a first-person initiative of the Brazilian scientific community to evaluate its own practices. We will also be impersonal in our choice of results to replicate, working with random samples and performing our analysis at the level of experiments; thus, even if a finding is not deemed reproducible, this will not necessarily invalidate an article’s conclusions or call a researcher into question.
An additional challenge is to ensure that participating labs have sufficient expertise with a methodology or model to provide accurate results. Ensuring that the original protocol is indeed being followed is likely to require steps such as cell line/animal strain authentication and positive controls for experimental validation. Nevertheless, we prefer this naturalistic approach to the alternative of providing each laboratory with animals or samples from a single source, which would inevitably underestimate variability. Moreover, while making sure that a lab is capable of performing a given experiment adequately is a challenge we cannot address perfectly, this is a problem of science as a whole – and if our project can build expertise on how to perform minimal certification of academic laboratories, this could be useful for other purposes as well.
A final challenge will be to put the results into perspective once they are obtained. Based on the results of previous reproducibility projects, a degree of irreproducibility is expected and may raise concerns about Brazilian science, as there will be no estimates from other countries for comparison. Nevertheless, our view is that, no matter the results, they are bound to put Brazil at the vanguard of the reproducibility debate, if only because we will likely be the first country to produce such an estimate.
Conclusions
With the rise in awareness over reproducibility issues, systematic replication initiatives have begun to develop in various research fields (Camerer et al., 2016; Camerer et al., 2018; Cova et al., 2018; Errington et al., 2014; Open Science Collaboration, 2015; Tan et al., 2015). Our study offers a different perspective on the concept, covering different research areas in the life sciences with focus in a particular country.
This kind of initiative inevitably causes controversy both on the validity of the effort (Coyne, 2016; Nature Medicine, 2016) and on the interpretation of the results (Baker and Dolgin, 2017; Gilbert et al., 2016; Patil et al., 2016). Nevertheless, multicenter replication efforts are as much about the process as about the data. Thus, if we attain enough visibility within the Brazilian scientific community, a large part of our mission – fostering the debate on reproducibility and how to evaluate it – will have been achieved. Moreover, it is healthy for scientists to be reminded that self-correction and confirmation are a part of science, and that published findings are passive of independent replication. There is still much work to be done in order for replication results to be incorporated into research assessment (Ioannidis, 2014; Munafò et al., 2017), but this kind of reminder by itself might conceivably be enough to initiate cultural and behavioral change.
Finally, for those involved as collaborators, one of the main returns will be the experience of tackling a large scientific question collectively in a transparent and rigorous way. We believe that large-scale efforts can help to lead an overly competitive culture back to the Mertonian ideal of communality, and hope to engage both collaborators and the Brazilian scientific community at large through data sharing, public consultations and social media (via our website: http://reprodutibilidade.bio.br/home). The life sciences community in Brazil is large enough to need this kind of challenge, but perhaps still small enough to answer cohesively. We thus hope that the Brazilian Reproducibility Initiative, through its process as much as through its results, can have a positive impact on the scientific culture of our country for years to come.
Data availability
All data cited in the article is available at the project's site at the Open Science Framework (https://osf.io/6av7k/).
-
Open Science FrameworkData from The Brazilian Reproducibility Initiative: a systematic assessment of Brazilian biomedical science.
References
-
WebsiteConsiderações sobre o processo de avaliação da pós-graduação da CAPESAccessed January 25, 2019.
-
Dez coisas que você deveria saber sobre o QualisRevista Brasileira De Pós-Graduação 13:13–40.https://doi.org/10.21713/2358-2332.2016.v13.947
-
Estimating the reproducibility of experimental philosophyReview of Philosophy and Psychology 28:1–36.https://doi.org/10.1007/s13164-018-0400-9
-
Many Labs 3: Evaluating participant pool quality across the academic semester via replicationJournal of Experimental Social Psychology 67:68–82.https://doi.org/10.1016/j.jesp.2015.10.012
-
What does research reproducibility mean?Science Translational Medicine 8:341ps12.https://doi.org/10.1126/scitranslmed.aaf5027
-
Sorting out the FACS: a devil in the detailsCell Reports 6:779–781.https://doi.org/10.1016/j.celrep.2014.02.021
-
Os planos nacionais de Pós-graduação (PNPG) e suas repercussões na pós-graduação brasileiraPerspectiva 24:133–160.
-
How to make more published research truePLOS Medicine 11:e1001747.https://doi.org/10.1371/journal.pmed.1001747
-
A manifesto for reproducible scienceNature Human Behaviour 1:0021.https://doi.org/10.1038/s41562-016-0021
-
What should researchers expect when they replicate studies? A statistical view of replicability in psychological sciencePerspectives on Psychological Science 11:539–544.https://doi.org/10.1177/1745691616646366
-
Believe it or not: how much can we rely on published data on potential drug targets?Nature Reviews Drug Discovery 10:712.https://doi.org/10.1038/nrd3439-c1
-
WebsiteBrasil cresce em produção científica, mas índice de qualidade caiFolha De S. Paulo. Accessed January 25, 2019.
-
WebsiteCarta aberta ao presidente da república em defesa da capes recebe mais de 50 assinaturas e é destaque na imprensa nacionalAccessed January 25, 2019.
-
WebsiteUm espaço para ciência: a formação da comunidade científica no brasilAccessed January 25, 2019.
-
Many analysts, one data set: Making transparent how variations in analytic choices affect resultsAdvances in Methods and Practices in Psychological Science 1:337–356.https://doi.org/10.1177/2515245917747646
-
Small telescopes: detectability and the evaluation of replication resultsPsychological Science 26:559–569.https://doi.org/10.1177/0956797614567341
-
WebsiteProstate Cancer Foundation-Movember Foundation Reproducibility InitiativeAccessed January 25, 2019.
Decision letter
-
Peter RodgersSenior and Reviewing Editor; eLife, United Kingdom
-
Timothy M ErringtonReviewer; Center for Open Science, United States
-
Richard KleinReviewer; Université Grenoble Alpes, France
In the interests of transparency, eLife includes the editorial decision letter and accompanying author responses. A lightly edited version of the letter sent to the authors after peer review is shown, indicating the most substantive concerns; minor comments are not usually included.
Thank you for submitting your article "The Brazilian Reproducibility Initiative: a systematic assessment of Brazilian biomedical science" to eLife for consideration as a Feature Article. Your article has been reviewed by two peer reviewers, and the evaluation has been overseen by the Peter Rodgers, eLife Features Editor. The following individuals involved in review of your submission have agreed to reveal their identity: Timothy M Errington (Reviewer #1); Richard Klein (Reviewer #2).
The reviewers have discussed the reviews with one another and the Reviewing Editor has drafted this decision to help you prepare a revised submission.
Summary:
There is a clear need for these sorts of projects to re-evaluate where we stand, across all scientific disciplines, in light of the replication crises already observed in psychology and other fields. This project seems well organized and thought-out, but could be strengthened by addressing the following comments.
Essential revisions:
a) The article needs to be as up-to-date as possible when published. For example, if the 3-5 methods have been identified, the article needs to say what they are and how they were chosen; if they have not been identified, the description of the selection process needs to be improved (see point j below). Likewise for the recruitment of collaborators.
b) I agree that narrowing the replications to a signal experiment (really a single main effect) is a clean way to measure reproducibility across studies. However, there are many 'nested' aspects to an experiment that need to be considered for proper interpretation. For example, testing the impact of depleting a particular gene of interest on some outcome (e.g. apoptosis) could require multiple experimental techniques (e.g. PCR or Western blot to confirm depletion, flow cytometry to detect apoptosis). Will all control related experimental aspects be included in the replication (in the example above PCR or Western blot) in addition to the key outcome (flow cytometry)? Also, it sounds as if just a single comparison (e.g. treatment vs control; https://osf.io/57f8s/) will be performed for each experiment: will positive and/or negative controls also be included?
c) Subsection “The Brazilian Reproducibility Initiative: aims and scope”, second paragraph. Please be more explicit about how generalizable your results will be to biomedical research in Brazil, Brazilian science in general, and biomedical research in general, given your sample size and biases imposed by your inclusion/exclusion criteria (e.g. use of commercially available materials (subsection “Selection of methods”, last paragraph).
d) Please also be more explicit about the caveats associated with having only a few labs if one goal of the project is to identify particular steps of a technique that are associated with a lack of reproducibility. There are numerous steps in a given technique and little variation with only 3 replications (as mentioned in the third paragraph of the subsection “Evaluating replications”). Nonetheless, the added benefit on the outcome by allowing for variation to occur will give a better understanding of the reproducibility (generalizability) of the finding across settings.
e) How much variation will be allowed? For example, if the original study states the species or sex of an animal, will that be held constant or will it be open to change among the labs? If a specific drug is stated, will that be kept constant or will it be open to change if it has the same MOA? Each lab should preregister experiments based on a common framework of what will not be changed (i.e. what they and the project leads agree are perceived necessary features of the original experiment). The worry I have is that if left open to interpretation too broadly, the three experiments that are replications will be more conceptual than direct and less comparable to each other or the original as much as the authors intend (see: Nosek and Errington, 2017; doi.org/10.7554/eLife.23383).
f) Two possible questions:
1) Can the findings be reproduced based on the published literature?
2) Can the findings be reproduced under ideal conditions?
Both are extremely important, but by blinding teams to feedback/contact with original authors you may be favoring #1 at some expense to #2. You might consider (but do not have to do this) experimentally testing for the influence of original author feedback by giving one of the three labs for each site any extra details provided by the original authors (and perhaps further ask them to review that lab's procedure). This could then both estimate reproducibility and inform which solutions to prioritize to improve it (e.g., improved documentation). No idea what response rate you'd expect, but in Many Labs 2 we got feedback from original authors or close collaborators in 28/28 studies.
g) I appreciate the difficulty defining replication success, but if the most important outcome is whether an effect is observed or not then I'm not sure the current primary definition is optimal. You may consider defining a 'smallest effect size of interest' and combining that with significance testing (e.g., r >.1 and p <.001).
h) It is not clear how the individual studies and the overall paper will be peer reviewed. Will the individual studies be handled in a similar way to the RP:CB? (That is, there is a Registered Report for each individual study that is reviewed and must be accepted before data collection can begin, and there is an agreement to publish the results, regardless of outcome, so long as the protocol in the Registered Report has been adhered to). And what process will be used to peer review the overall paper? I highly recommend a structured procedure to ensure quality and combat any possible publication bias against null findings. For most Many Lab projects we've included original authors as part of this review process, but I understand the arguments against this.
i) Please include a flow chart (such as a PRISMA flow diagram) to show how the papers to be replicated will be selected. I also have some questions about the selection process as described in the present manuscript. The text states that 5000 articles were assessed (subsection “Selection of methods”, second paragraph), but the table legend mentions an initial survey of 100 articles (is that 100 of the 5000?). Also, if I count the number of occurrences of each technique among the main experiments, I count 51, not 100, which suggests 49 are being excluded for some reason. Please also consider including a figure along the lines of Figures 3-5 in https://osf.io/qhyae/ to show the range of techniques used.
j) Subsection “Selection of methods”, second paragraph: How will the list of 10 commonly used techniques (shown in Table 1) be narrowed down to 3-5 methods? Will cost be a factor? Also, will the distribution of biological models used in the replications match the overall distribution (i.e., rodents > cell lines > micro-organisms etc.), or will simple randomization occur? And likewise, for the techniques used?
https://doi.org/10.7554/eLife.41602.008Author response
Essential revisions:
a) The article needs to be as up-to-date as possible when published. For example, if the 3-5 methods have been identified, the article needs to say what they are and how they were chosen; if they have not been identified, the description of the selection process needs to be improved (see point j below). Likewise for the recruitment of collaborators.
We naturally agree with the point of updating the manuscript with the current state of the project. In this sense, the revision process has allowed some important steps to be concluded. Over 3 months of registration, we’ve had 71 laboratories across 19 states of Brazil sign up as potential collaborators of the initiative.
On the basis of this network (as well as of methodological and budget concerns, as will be described below), we have selected five methods to be included in the replication effort: namely, MTT, RT-PCR, elevated plus maze, western blot and immunohistochemistry. We will start the replication experiments with three of these (MTT, RT-PCR and elevated plus maze), totaling 60 experiments, and will add the other two after full protocol development for the first three, as this will allow us to have a clearer estimation of the project’s workload and costs.
These details have now been updated in the third paragraph of the subsection “Selection of methods”, and in other points of the manuscript. The selection process has also been described in clearer detail in the flowchart presented in Figure 1C, as suggested below, which has replaced Table 1 of the original manuscript.
b) I agree that narrowing the replications to a signal experiment (really a single main effect) is a clean way to measure reproducibility across studies. However, there are many 'nested' aspects to an experiment that need to be considered for proper interpretation. For example, testing the impact of depleting a particular gene of interest on some outcome (e.g. apoptosis) could require multiple experimental techniques (e.g. PCR or Western blot to confirm depletion, flow cytometry to detect apoptosis). Will all control related experimental aspects be included in the replication (in the example above PCR or Western blot) in addition to the key outcome (flow cytometry)? Also, it sounds as if just a single comparison (e.g. treatment vs control; https://osf.io/57f8s/) will be performed for each experiment: will positive and/or negative controls also be included?
The review points out an important point that we agree was not completely clear in the original manuscript. Our article/experiment selection will be based on the main technique of interest – i.e. that used to measure the outcome variable. That said, as the whole experiment will be replicated, there are naturally other methods that will be involved in performing the required experimental interventions, and eventually for adding necessary controls.
In our recruitment process, we asked for reasonably detailed information on the expertise of the participating laboratories – not only in the main techniques of the project, but also in handling biological models, performing interventions, dissecting tissue, etc. Thus, one of the criteria for inclusion of articles will be that we have at least three participating labs with the required expertise to perform the whole experiment. This will be confirmed with the replicating labs after screening to confirm inclusion of the experiment. If a given experiment requires expertise that is not available in at least three labs, it will not be included in the replication initiative. This is now explained in the fourth and last paragraphs of the subsection “Selection of methods”.
Our main result of interest will be indeed based on a single experiment – i.e. a comparison in a dependent variable between two groups – in order to facilitate statistical analysis for the whole sample. Nevertheless, the reviewers are right in pointing out that, in some experiments, additional controls might be necessary for interpreting the results – e.g. positive controls to confirm the sensitivity of the detection method, for example. If such controls are part of the original experiment, they will be included in the replication as well, unless for some reason this is not technically feasible. If not, the replicating teams will still be allowed to suggest controls when they are judged necessary. The need for these controls will be reviewed during the protocol revision process – see response to point (h) below – in order to confirm inclusion, as explained in the seventh paragraph of the subsection “Multicenter replication”.
c) Subsection “The Brazilian Reproducibility Initiative: aims and scope”, second paragraph. Please be more explicit about how generalizable your results will be to biomedical research in Brazil, Brazilian science in general, and biomedical research in general, given your sample size and biases imposed by your inclusion/exclusion criteria (e.g. use of commercially available materials (subsection “Selection of methods”, last paragraph).
Generalizability of the results will certainly be limited by many factors, including the selected techniques/biological models and our inclusion/exclusion criteria for selecting experiments – which include cost, commercial availability and expertise of the replicating labs. This is now made clear not only in the above-mentioned passage (subsection “The Brazilian Reproducibility Initiative: aims and scope”, second paragraph) but also in the sixth paragraph of the subsection “Evaluating replications”.
d) Please also be more explicit about the caveats associated with having only a few labs if one goal of the project is to identify particular steps of a technique that are associated with a lack of reproducibility. There are numerous steps in a given technique and little variation with only 3 replications (as mentioned in the third paragraph of the subsection “Evaluating replications”). Nonetheless, the added benefit on the outcome by allowing for variation to occur will give a better understanding of the reproducibility (generalizability) of the finding across settings.
The reviewers are right in pointing out that our ability to detect variation due to any particular steps of the selected techniques will be limited by the number of replicating labs and by the amount of variation between protocols – which cannot be predicted in advance, as it will depend on how different labs will interpret and adapt the published protocols. Nevertheless, as our primary goal is to examine the reproducibility of the published literature in view of naturalistic interlaboratory variability, this is a necessary compromise: for laboratories to be free in making their own choices, we must refrain from controlling variation.
In view of this, we are fully aware both that (a) investigation of the effect of individual steps on variability will be a secondary analysis which will be limited in terms of scope and statistical power and that (b) it is hard to predict in advance how limited this scope will be. For this reason, we will work on the analysis plan for this part of the project only after protocols have been built (but before the experiments are performed), in order to have a better idea of the range of methodological variability. More likely than not, we will have to substitute analysis of individual steps of the methods for large-scale, quantitative estimates of variability across the whole experiment or its general sections.
That said, we point out that, although each experiment will be performed only in three labs, we will have 20 experiments with each technique with the same methodological steps described. Thus, even if in a single experiment it may be impossible to pinpoint sorts of variation, on the aggregate we might have some idea on what sources of methodological variability correlate more strongly with variation in results.
An in-depth discussion of these analysis options seems premature at this point of the project – and is certainly beyond the scope of the current manuscript. Nevertheless, we now acknowledge the above-mentioned limitations more clearly in the fourth paragraph of the subsection “Evaluating replications”.
e) How much variation will be allowed? For example, if the original study states the species or sex of an animal, will that be held constant or will it be open to change among the labs? If a specific drug is stated, will that be kept constant or will it be open to change if it has the same MOA? Each lab should preregister experiments based on a common framework of what will not be changed (i.e. what they and the project leads agree are perceived necessary features of the original experiment). The worry I have is that if left open to interpretation too broadly, the three experiments that are replications will be more conceptual than direct and less comparable to each other or the original as much as the authors intend (see: Nosek and Errington, 2017; doi.org/10.7554/eLife.23383).
As stated in the manuscript (subsection “Multicenter replication”, fifth paragraph), replications will be as direct as possible in the sense of explicitly following the protocol described in the original manuscript. Naturally, there will be cases in which adaptations are required – e.g. use of different equipment or reagent when the original is not available. Nevertheless, all these adaptations will be revised by the coordinating team and an independent laboratory (see protocol review process below) so that it is agreed that the adapted protocol maintains the necessary features of the original experiment.
On the other hand, steps that are not described in the original protocol will be left to vary as much as possible. This is necessary to approach our main goal of measuring naturalistic reproducibility – i.e. question 1 in point (f) below. Although these steps will be left to vary, we will record these protocol variations in as much details as possible using standardized forms, in order to allow for the analyses mentioned in the point above.
An exception to the rule of following the original protocol as closely as possible will be if we feel that additional steps are necessary to ensure that the experiments are performed free of bias – for example, unblinded experiments will be blinded and non-randomized experiments will be randomized whenever possible. As described above, we will also add necessary controls when deemed necessary (even though the main experiment is left unchanged). This is now explained in more detail in the seventh paragraph of the subsection “Multicenter replication”.
f) Two possible questions:
1) Can the findings be reproduced based on the published literature?
2) Can the findings be reproduced under ideal conditions?
Both are extremely important, but by blinding teams to feedback/contact with original authors you may be favoring #1 at some expense to #2. You might consider (but do not have to do this) experimentally testing for the influence of original author feedback by giving one of the three labs for each site any extra details provided by the original authors (and perhaps further ask them to review that lab's procedure). This could then both estimate reproducibility and inform which solutions to prioritize to improve it (e.g., improved documentation). No idea what response rate you'd expect, but in Many Labs 2 we got feedback from original authors or close collaborators in 28/28 studies.
The reviewers are correct in stating that these two questions are different – and perhaps mutually exclusive. Our option from the start of the project has always been to answer question (1) – i.e. reproducibility in naturalistic conditions upon reading the literature, and we are indeed favoring it deliberately. This has been made clear in the fifth paragraph of the subsection “Multicenter replication”.
The suggestion to try to address both questions at the same time is certainly interesting – however, we feel that it could complicate the analysis of the results. As some replications will be more alike to the original experiment than the others, our estimate of interlaboratory variability would be slightly more biased towards the original experiment than what would be expected in naturalistic replication. We can solve this by removing these labs from the analysis, but that would leave only 2 laboratories performing truly independent, unguided replications.
Another problem with this approach is that, even if we do get feedback from most original authors – something on which we are not clearly counting on – we will be replicating experiments published over 20 years (e.g. between 1998 and 2017), which means that much protocol information will likely have been lost for older experiments. Moreover, responder bias among authors could also cause information to be more available for particular types of authors or institutions; thus, using author information only when it’s available would likely bias our analyses concerning predictors of reproducibility.
With this in mind, although we find the suggestion interesting, we would rather keep up with our original plot of focusing on naturalistic reproducibility based only on published information, as explained inthe sixth paragraph of the subsection “Multicenter replication”.
g) I appreciate the difficulty defining replication success, but if the most important outcome is whether an effect is observed or not then I'm not sure the current primary definition is optimal. You may consider defining a 'smallest effect size of interest' and combining that with significance testing (e.g., r >.1 and p <.001).
We also appreciate the difficulty in defining replication success – and have opted for multiple measures, as previous replication initiatives have done, precisely because of that. That said, we are somewhat skeptical of defining a ‘smallest effect size of interest’, as ‘typical’ effect sizes seem to vary a lot across areas of science depending on the type of interventions that are used. Our own work in rodent fear conditioning, for example (Carneiro et al., 2018) shows that the arbitrary criteria for ‘small’, ‘moderate’ and ‘large’ effects commonly used in psychology do not hold at all in that context. Moreover, the importance of effect sizes is unfortunately neglected in most fields of biology (again, see Carneiro et al., 2018 for an example). Thus, as we will be looking across different areas of science and interventions, we do not feel confident in defining a smallest effect size of interest.
The statistical significance of the pooled replication studies is already one of our definitions of a successful replication, as has been the case in most large replication initiatives. Nevertheless, we would prefer not to be so stringent as to lower the significance threshold to p < 0.001. In keeping in line with other replication studies, and with most of the biomedical literature, we would prefer to keep up with the standard p < 0.05. We fully understand that this means that some false positives will be expected within the replication effort, but we feel maintaining the declared false-positive rate of the original studies provides a fairer assessment of whether these studies might have contained unjustified claims. Nevertheless, we commit to reporting exact p values for all experiments, in order to allow our data to be interpreted under different threshold assumptions.
h) It is not clear how the individual studies and the overall paper will be peer reviewed. Will the individual studies be handled in a similar way to the RP:CB? (That is, there is a Registered Report for each individual study that is reviewed and must be accepted before data collection can begin, and there is an agreement to publish the results, regardless of outcome, so long as the protocol in the Registered Report has been adhered to). And what process will be used to peer review the overall paper? I highly recommend a structured procedure to ensure quality and combat any possible publication bias against null findings. For most Many Lab projects we've included original authors as part of this review process, but I understand the arguments against this.
We thank the reviewers for bringing the point about peer review of the protocols, which we had not addressed in the original manuscript – partly because we had not discussed it in sufficient depth at the time of submission. We certainly do not plan to publish each experiment as a peer-reviewed Registered Report, as the individual experiments by themselves (unlike the larger studies included in RP:CB) would probably be of little interest to readers. That said, we agree on the importance of peer review happening before experiments are performed.
For this, we will use a similar approach to that used in the recent “Many Analysts, One Dataset” project (Silberzahn et al., 2018) and use our own base of collaborating labs – which we now know to be large enough – to provide peer review on the first version of the protocols. Each protocol will be reviewed both by a member of the coordinating team and by an independent laboratory participating in the Initiative with expertise in the same technique. These reviewers will be instructed to (a) verify whether there are adaptations to the originally described protocol and adjudicate whether any of them constitute a deviation that is large enough to make it invalid as a direct replication and (b) verify whether necessary measures and controls to reduce risk of bias and ensure the validity of results were taken. After peer review, each laboratory will receive feedback from both reviewers and will be allowed to revise the protocol, which will be reviewed once more by the same reviewers for approval before the start of the experiments. This is now discussed in the last paragraph of the subsection “Multicenter replication”.
We will review each protocol independently – i.e. each of the 3 different versions of an experiment will be reviewed by a different reviewer – in order to minimize the possibility of ‘overstandardization’ – i.e. to prevent that having a single reviewer revising all the protocols would make them more similar than would be expected from independent replication. In keeping up with the idea of naturalistic replication, as detailed above – as well as to preserve blinding – we will not include feedback from the original authors as part of the review – although we also understand the argument in favor of this. We discuss all of these issues in the aforementioned subsection.
Concerning the publication plan itself, our current view is that the actual article describing the whole set of results will be submitted as a regular paper after completion of the experiments – even though all protocols will have been reviewed and preregistered at the Open Science Framework in advance. We also envision that the results within each technique might deserve a separate, in-depth article examining sources of variability within each kind of experiment and their relationship to reproducibility. Finally, it is possible that some of the secondary analyses – such as researcher predictions and correlates of reproducibility, could generate other articles, although we would rather keep these in the main publication. Such concerns, however, are probably premature at the moment, and the ideal form of publication will become clearer as the project develops.
i) Please include a flow chart (such as a PRISMA flow diagram) to show how the papers to be replicated will be selected. I also have some questions about the selection process as described in the present manuscript. The text states that 5000 articles were assessed (subsection “Selection of methods”, second paragraph), but the table legend mentions an initial survey of 100 articles (is that 100 of the 5000?). Also, if I count the number of occurrences of each technique among the main experiments, I count 51, not 100, which suggests 49 are being excluded for some reason. Please also consider including a figure along the lines of Figures 3-5 in https://osf.io/qhyae/ to show the range of techniques used.
We thank the reviewer for the excellent suggestion concerning the flow chart for selection of experiments. It has now been included as Figure 1C. That said, experiment selection will continue over the next months, with the groups of participating labs already defined, in order to arrive at a sample of experiments that is within our available expertise.
The initial survey was performed separately from the article selection process, as in the former our goal was to assess candidate techniques widely performed within the country. For this reason, in this step we only worked with articles in which all authors were located in Brazil, in order to make sure that the methods were available locally. The main results of the survey are now included in Figure 1A and 1B. A wide range of methods was detected in this step, and the figure only shows the most common results; thus, the results shown in the bar graphs are non-extensive and do not add up to 100.
j) Subsection “Selection of methods”, second paragraph: How will the list of 10 commonly used techniques (shown in Table 1) be narrowed down to 3-5 methods? Will cost be a factor? Also, will the distribution of biological models used in the replications match the overall distribution (i.e., rodents > cell lines > micro-organisms etc.), or will simple randomization occur? And likewise, for the techniques used?
Our list of candidate techniques has now been narrowed down to 5, as explained in the third paragraph of the subsection “Selection of methods” and in Figure 1C. The criteria used to define these techniques were (a) the number of labs with expertise in this methods that signed up for the initiative, (b) the availability of experiments in an initial screening sample that matched the labs’ expertise while remaining under our cost limit and (c) the total number of labs that could be included with the particular combination chosen. Thus, even though fewer laboratories signed up as candidates for behavioral techniques, the elevated plus maze was included due to its low cost and to the availability of a pool of laboratories that was largely independent from those signed up for the molecular techniques (e.g. MTT, RT-PCR, western blot).
Concerning the number of experiments included, our aim is to include 20 experiments per technique – thus, the distribution of techniques is set a priori and will not match their actual prevalence within the literature. Within each technique, however, the biological models will not be constrained – thus, the prevalence of experiments using rodents and cell lines should approach that observed in the literature for each method, although this prevalence could be biased by cost or expertise issues. This is now better explained in the fifth paragraph of the subsection “Selection of methods”.
https://doi.org/10.7554/eLife.41602.009Article and author information
Author details
Funding
Instituto Serrapilheira
- Olavo B Amaral
Conselho Nacional de Desenvolvimento Científico e Tecnológico
- Clarissa FD Carneiro
The project's funder (Instituto Serrapilheira) made suggestions on the study design, but had no role in data collection and interpretation, or in the decision to submit the work for publication. KN and APWS are supported by post-doctoral scholarships within this project. CFDC is supported by a PhD scholarship from CNPq.
Publication history
- Received:
- Accepted:
- Accepted Manuscript published:
- Version of Record published:
Copyright
© 2019, Amaral et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,900
- views
-
- 255
- downloads
-
- 31
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 31
- citations for umbrella DOI https://doi.org/10.7554/eLife.41602






