Natural selection and repeated patterns of molecular evolution following allopatric divergence
Figures
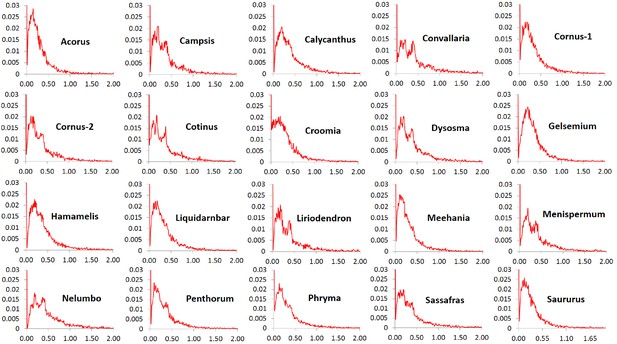
Frequency distribution of synonymous substitutions per synonymous site (Ks) of putative orthologs (POGs) from leaf transcriptomes of 20 species/variety pairs of angiosperms.
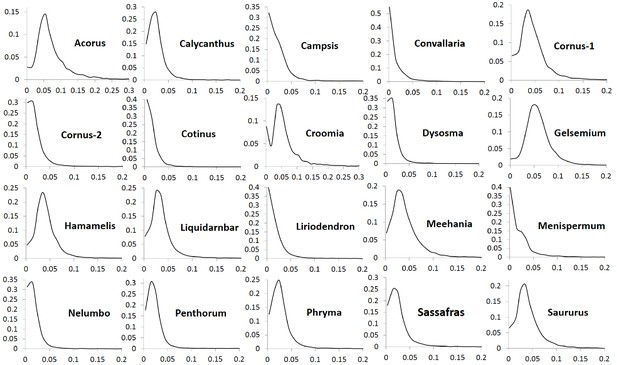
Frequency distribution of nonsynonymous substitutions per non-sysnonymous site (Ka) of putative orthologs (POGs) of leaf transcriptomes from 20 species/variety pairs of angiosperms.
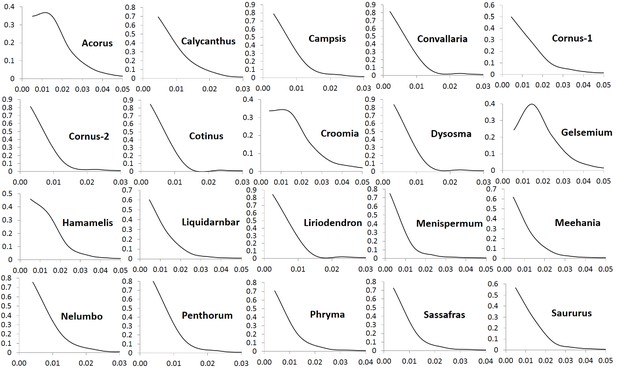
Frequency distribution of Ka/Ks ratios of putative orthologs (POGs) from leaf transcriptomes of 20 species/variety pairs of angiosperms.
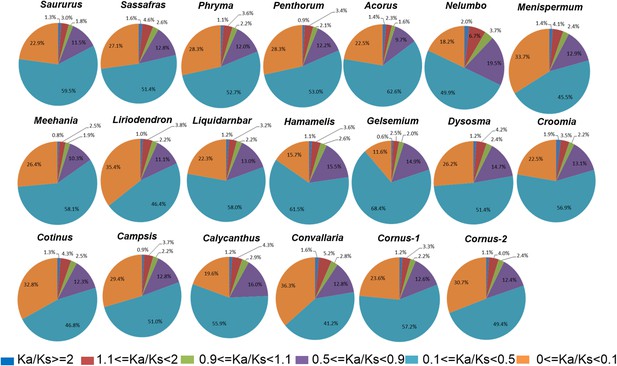
Relative abundance of putative orthologs (POGs) with different categories of Ka/Ks values.
Numbers of genes in each category are provided in Supplementary file 3.
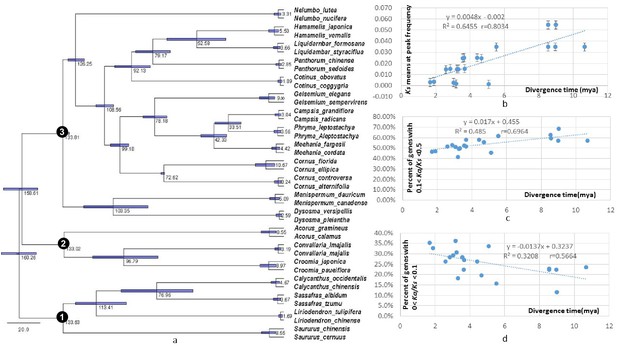
Dated global phylogeny of the 20 taxon pairs and correlations of divergence time with level of divergence at sysnonymous sites, withabundance of genes under moderate purifying selection (Ka/Ks = 0.1 - 0.5), and with abundance of genes under strong purifying selection (ka/Ks <0.1).
(a) Dated global phylogeny and divergence times of taxon pairs estimated using BEAST program and seven single-copy orthologs shared by all 40 taxa with no missing data. Number 1, 2, and 3 indicate the crown node of Magnoliidae, crown node of Monocotyledoneae, and crown node of Eudicotyledoneae, respectively. (b – d). Correlation between divergence time and (b) Ks value at peak frequency of each genus, (c) abundance of POGs with Ka/Ks values between 0.1–0.5, and (d) abundance of POGs with Ka/Ks values < 0.1. Data used for the analyses are available in Supplementary file 4.
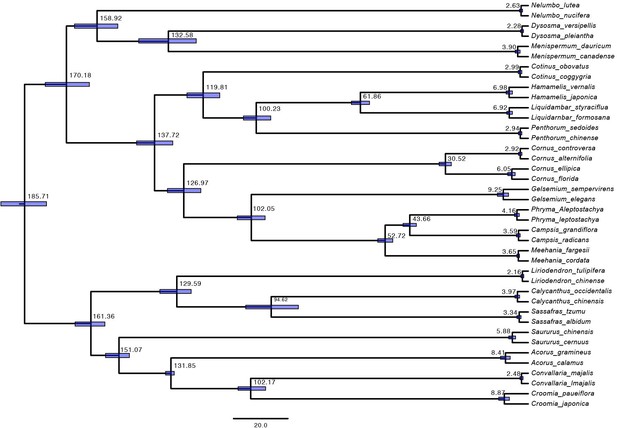
Divergence times of 20 taxon pairs estimated with 79 single copy orthologs present in 90% or more species using BEAST.
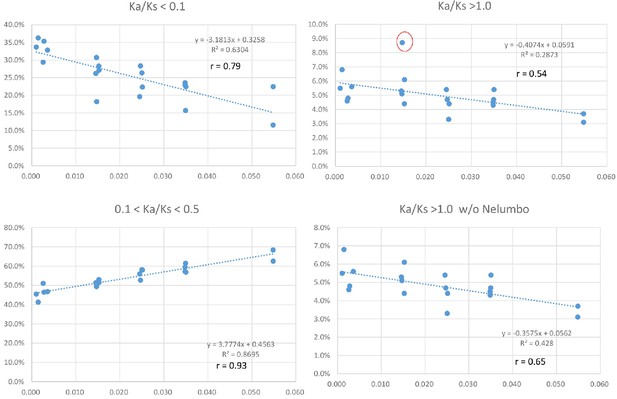
Positive and negative relationships between relative abundance (Y-axis) of genes with Ka/Ks values in the indicated ranges and Ks values of peak abundance (X-axis) in the 20 taxon pairs.
The pattern remains when the modified Ks (see Materials and methods) is used. Data used for the analyses are available in Supplementary file 4.
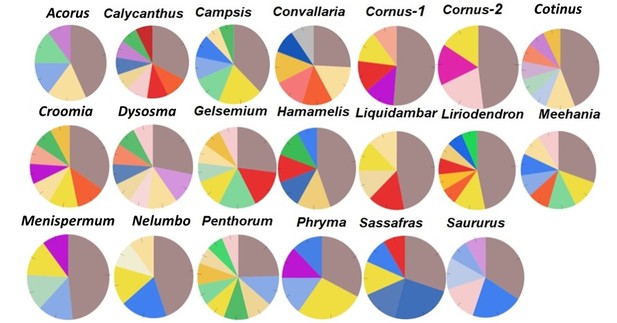
Relative abundance of genes with Ka/Ks > 2 annotated to Cell Component category, drawn from data in Supplementary file 5. Genes annotated to integral component of membrane (ICM) are shown in dark brown.
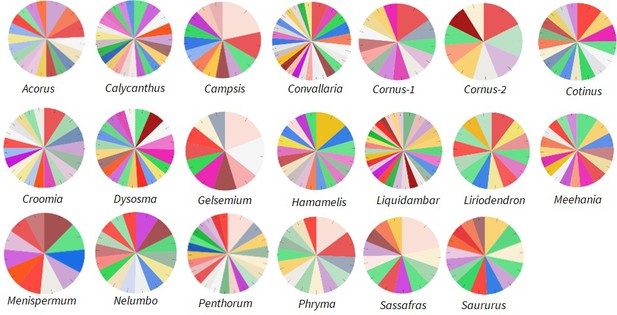
Relative abundance of Ka/Ks > 2 genes annotated to different functions in the Biological Process category; drawn from data in Supplementary file 5.
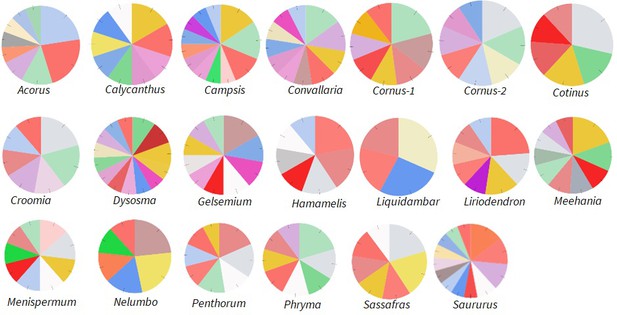
Relative abundance of Ka/Ks > 2 genes annotated to different functions of in the Molecular Function category; drawn from data in Supplementary file 5.
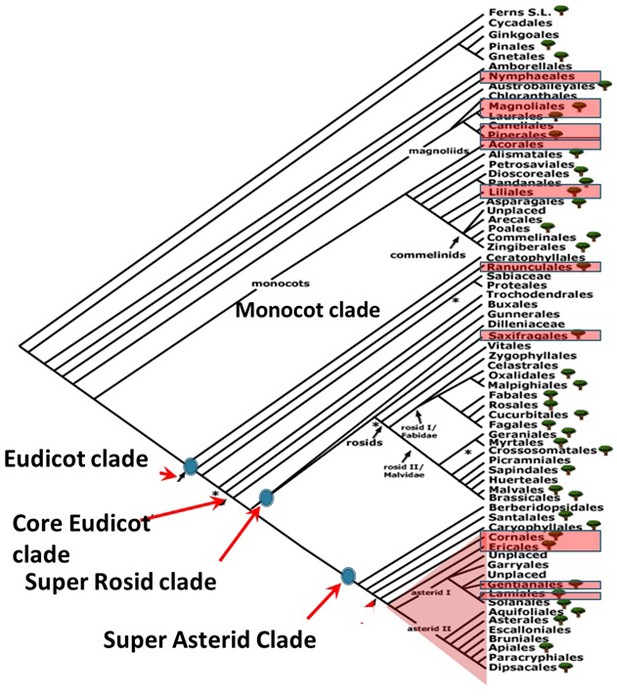
Phylogenetic positions of orders represented by species pairs sampled in the angiosperm phylogeny.
Phylogenetic tree was taken from Angiosperm phylogeny website.
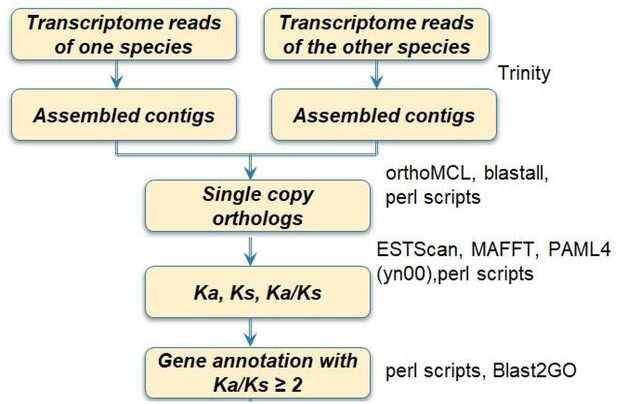
Flowchart of transcriptome sequence analyses for one species pair.
The process was repeated for 20 pairs. A customized pipeline of programs was developed for running these steps.
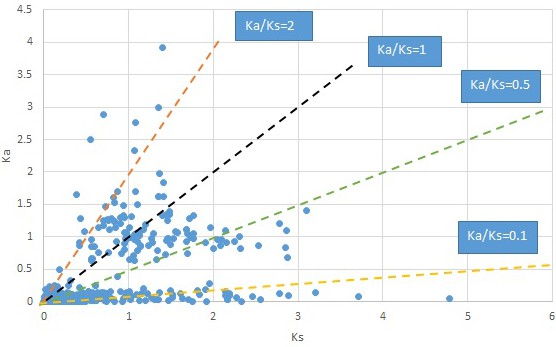
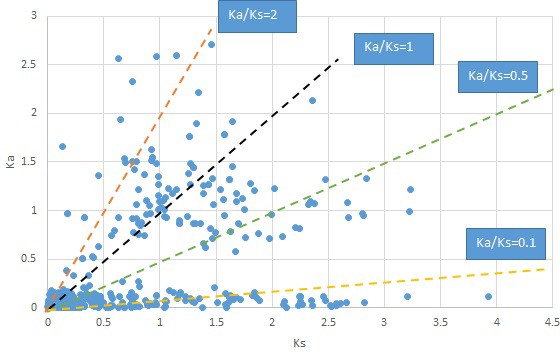
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Acorus.
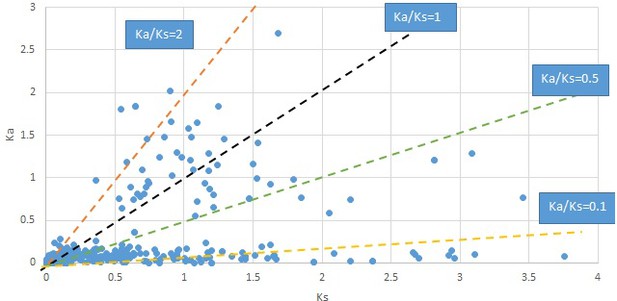
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Calycanthus.
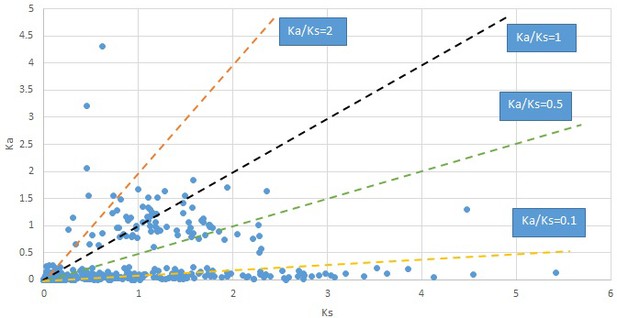
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Campsis.
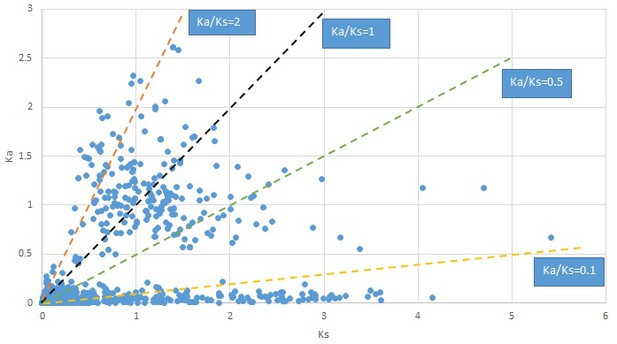
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Convallaria.
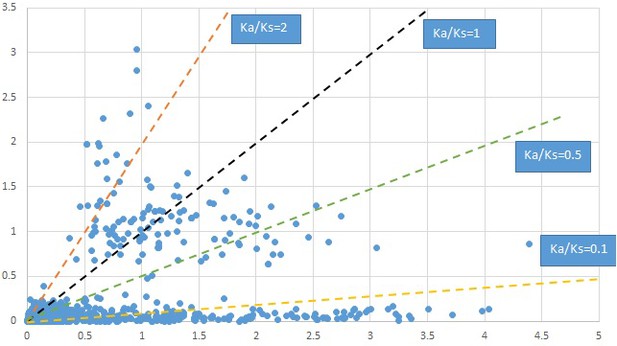
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Cornus-1.
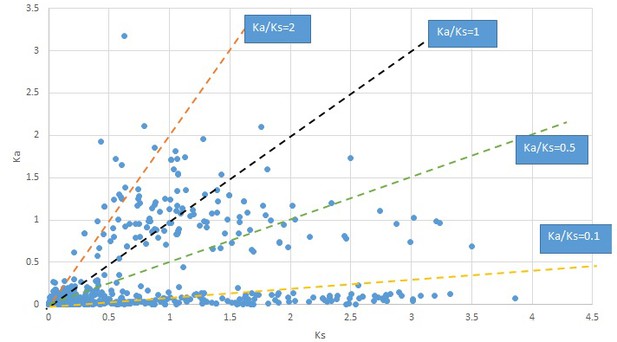
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Cornus-2.
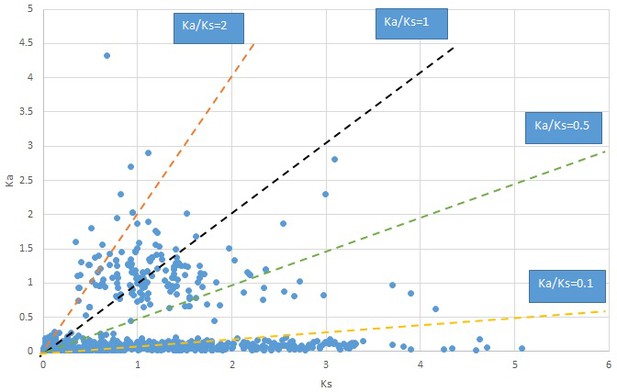
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Cotinus.
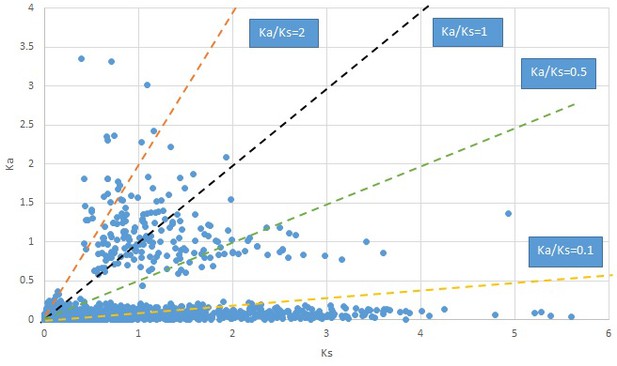
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Croomia.
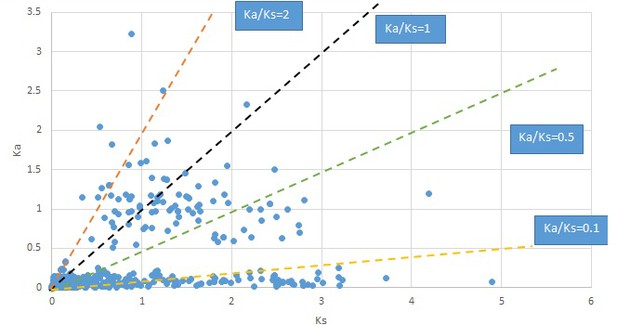
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Dysosma.
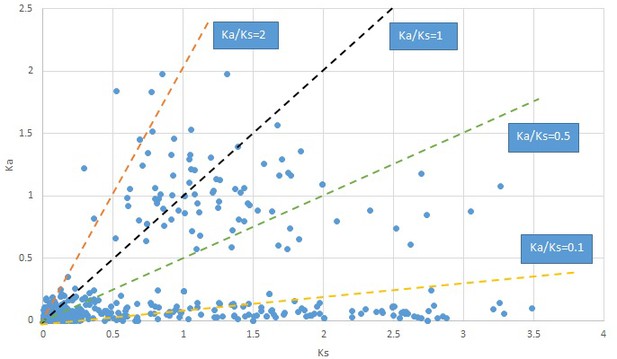
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Gelsemium.
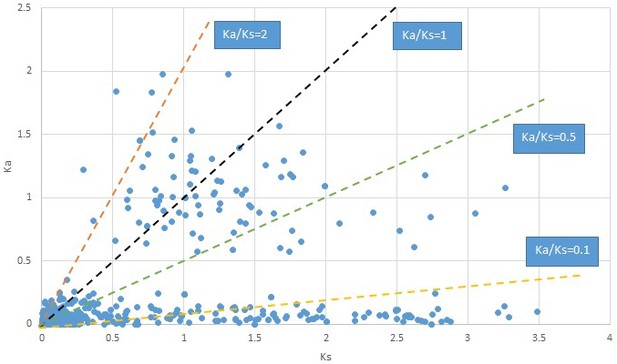
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Hamamelis.
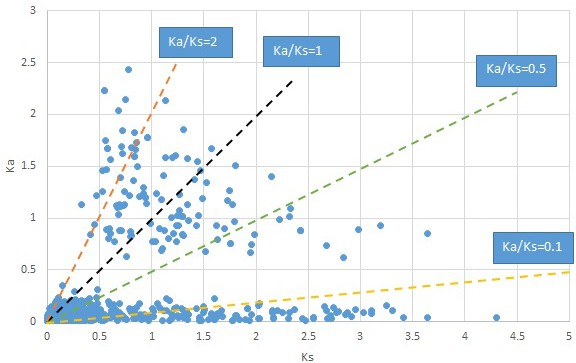
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Liquidarnbar.
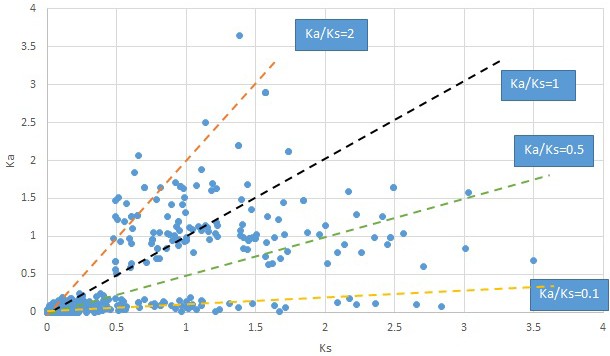
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Liriodendron.
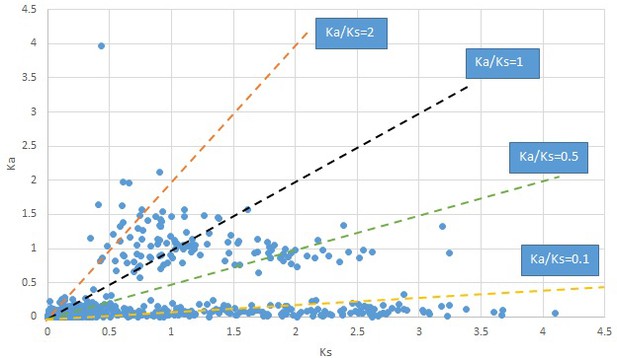
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Meehania.
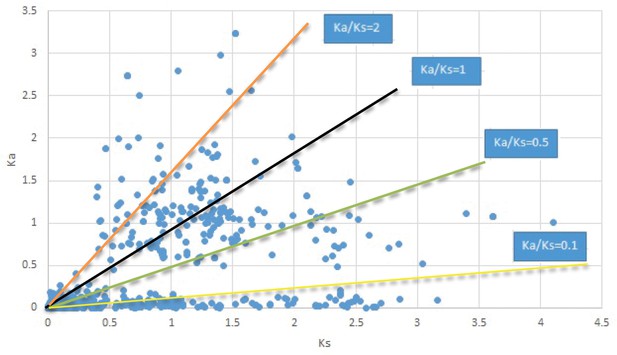
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Menispermum.
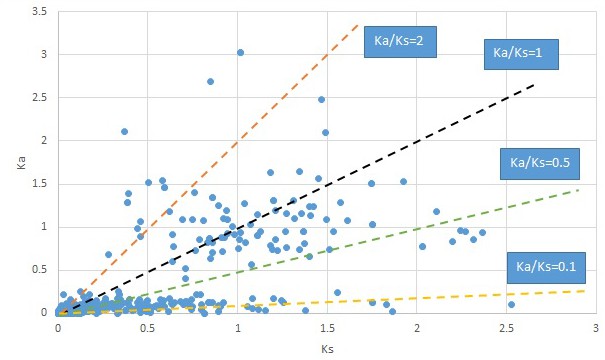
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Nelumbo.
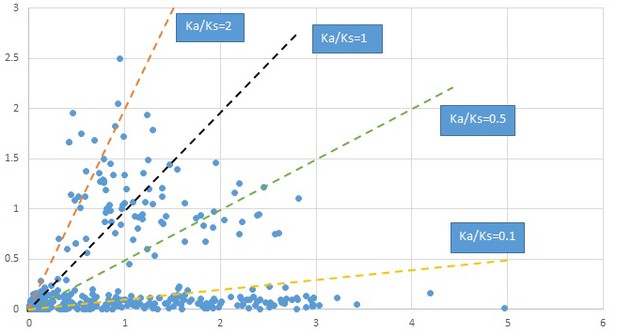
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Penthorum.
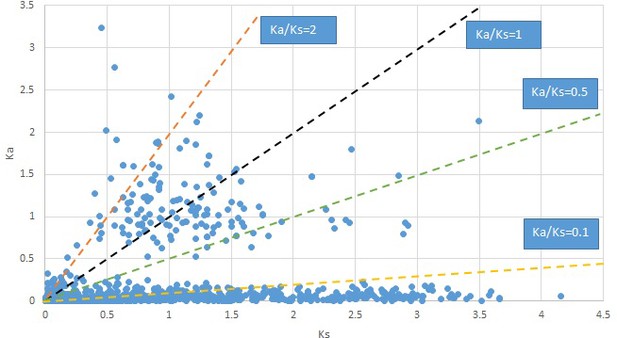
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Phryma.
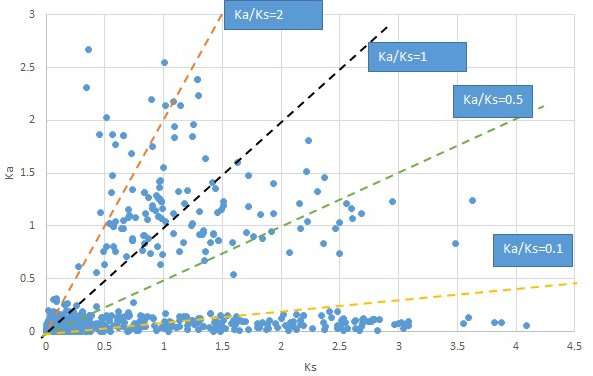
Plot of Ka and Ks values for each POG in 20 taxon pairs, in alphabetical order of genus names.
Sassafras.
Tables
List of biogeographic pairs and number of examined putative single copy orthologs (POGs) (sum from two species), maximum length (MAL), minimum length (MIL), and average length (AL) of POGs, number of POGs mapped to single copy genes in seed plants (NSP) and number of POGs that were mapped to single copy genes in 20 angiosperm plants (NAP).
| Genera | Species | Distribution | #of POGs | MAL of POGs (bp) | MIL of POGs (bp) | AL of POGs (bp) | NSP of POGs | NAP of POGs |
|---|---|---|---|---|---|---|---|---|
| Acorus | A.calamus, A.gramineus | EA-Asia/America | 17192/2 | 6213 | 102 | 874 | 77 | 81 |
| Calycanthus | C.chinensis, C.occidentalis | EA-WNA | 23116/2 | 6705 | 102 | 908 | 96 | 152 |
| Campsis | C.grandiflora, C.radicans | EA-ENA | 20840/2 | 5697 | 102 | 734 | 66 | 112 |
| Convallaria | C.majalis var. keiskei, C.majalisvar. Montana | EA-ENA | 30322/2 | 7041 | 102 | 502 | 53 | 75 |
| Cornus-1 | C.ellipica, C.florida | EA-ENA | 24398/2 | 5682 | 102 | 800 | 154 | 259 |
| Cornus-2 | C.alternifolia, C.controversa | EA-ENA | 26490/2 | 7455 | 102 | 836 | 210 | 323 |
| Cotinus | C.coggygria, C.obovatus | Eurasia-ENA | 26494/2 | 7374 | 102 | 889 | 237 | 361 |
| Croomia | C.japonica,C.paueiflora | EA-ENA | 23160/2 | 6480 | 102 | 817 | 71 | 112 |
| Dysosma | D.pleiantha, D.versipellis | Asia* | 24270/2 | 11694 | 102 | 944 | 132 | 218 |
| Gelsemium | G.elegans, G.sempervirens | EA-ENA | 22774/2 | 6801 | 102 | 980 | 129 | 194 |
| Hamamelis | H.japonica, H.vernalis | EA-ENA | 23052/2 | 7308 | 102 | 955 | 221 | 365 |
| Liquidarnbar | L.styraciflua, L.formosana | EA-ENA | 24844/2 | 7041 | 102 | 893 | 196 | 343 |
| Liriodendron | L.chinense, L.tulipifera | EA-ENA | 23060/2 | 7053 | 102 | 748 | 77 | 120 |
| Meehania | M.fargesii, M.cordata | EA-ENA | 25622/2 | 5562 | 102 | 919 | 97 | 166 |
| Menispermum | M.canadense, M.dauricum | EA-ENA | 27490/2 | 7347 | 102 | 449 | 60 | 121 |
| Nelumbo | N.lutea, N.nucifera | EA-ENA | 23988/2 | 7329 | 102 | 774 | 138 | 220 |
| Penthorum | P.chinense, P.sedoides | EA-ENA | 23288/2 | 15225 | 102 | 882 | 161 | 245 |
| Phryma | P.aleptostachya, P.leptostachya | EA-ENA | 25096/2 | 11445 | 102 | 863 | 106 | 180 |
| Sassafras | S.albidum, S.tzumu | EA-ENA | 25070/2 | 7524 | 102 | 809 | 104 | 153 |
| Saururus | S.cernuus, S.chinensis | EA-ENA | 19386/2 | 6300 | 102 | 888 | 53 | 84 |
-
*Evergreen-deciduous forests pair.
The number of POGs under strong positive selection (Ka/Ks > 2) in each taxon pair and results from Blast Search.
| Genera | Total | With blast (without hits) | With blast hits | With mapping | With GO annotation |
|---|---|---|---|---|---|
| Acorus | 101 | 18 | 20 | 17 | 46 |
| Calycanthus | 115 | 14 | 20 | 25 | 56 |
| Campsis | 72 | 5 | 15 | 10 | 42 |
| Convallaria | 123 | 51 | 12 | 15 | 45 |
| Cornus-1 | 123 | 17 | 16 | 23 | 67 |
| Cornus-2 | 116 | 7 | 12 | 29 | 68 |
| Cotinus | 124 | 40 | 11 | 13 | 60 |
| Croomia | 170 | 32 | 29 | 20 | 89 |
| Dysosma | 108 | 36 | 14 | 18 | 40 |
| Gelsemium | 67 | 5 | 10 | 8 | 44 |
| Hamamelis | 114 | 28 | 15 | 26 | 45 |
| Liquidambar | 122 | 23 | 19 | 21 | 59 |
| Liriodendron | 78 | 15 | 7 | 19 | 37 |
| Meehania | 85 | 12 | 14 | 7 | 52 |
| Menispermum | 106 | 18 | 6 | 19 | 63 |
| Nelumbo | 184 | 9 | 32 | 39 | 104 |
| Penthorum | 93 | 5 | 16 | 9 | 63 |
| Phryma | 119 | 4 | 24 | 16 | 75 |
| Sassafras | 159 | 24 | 27 | 31 | 77 |
| Saururus | 102 | 7 | 21 | 16 | 58 |
Number of genes with Ka/Ks > 2 ratios annotated to Integral Component of Membrane.
| Genera | Integral component of membrane |
|---|---|
| Acorus | 12 |
| Calycanthus | 13 |
| Campsis | 12 |
| Convallaria | 8 |
| Cornus-1 | 20 |
| Cornus-2 | 12 |
| Cotinus | 18 |
| Croomia | 26 |
| Dysosma | 7 |
| Gelsemium | 7 |
| Hamamelis | 16 |
| Liquidarnbar | 15 |
| Liriodendron | 7 |
| Meehania | 10 |
| Menispermum | 14 |
| Nelumbo | 26 |
| Penthorum | 15 |
| Phryma | 13 |
| Sassafras | 21 |
| Saururus | 16 |
Additional files
-
Supplementary file 1
Total number of orthologous gene families, number of putative single copy orthologous gene pairs (POGs) in each taxon pair, and the proportion of POGs.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp1-v4.xlsx
-
Supplementary file 2
Frequency of Ks values in 0.01 intervals of all pairs.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp2-v4.xlsx
-
Supplementary file 3
Number and relative abundance in percentage of POGs with Ka/Ks values distributed in different interval categories.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp3-v4.docx
-
Supplementary file 4
Information of divergence time estimated from BEAST using seven SCG genes shared by all 40 taxa on a global phylogeny, Ks value in peak frequency, and relative abundance of POGs in three Ka/Ks ratio ranges used in correlation analyses shown in Figures 5 and 6.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp4-v4.xlsx
-
Supplementary file 5
Results of gene annotation for genes with Ka/Ks > 2.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp5-v4.xlsx
-
Supplementary file 6
Results from Variation Cluster analysis of POGs with Ka/Ks > 2.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp6-v4.xls
-
Supplementary file 7
Data and results from scatter plot between divergence time and Ka/Ks ratio of 79 POGs shared by 90% or more taxon pairs.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp7-v4.xlsx
-
Supplementary file 8
Data and results for scatter plot of divergence time and Ka/Ks ratio (with Ka/Ks > 0.5 data points removed) of 79 POGs shared by 90% or more taxon pairs.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp8-v4.xlsx
-
Supplementary file 9
Functions of genes annotated as integral_compotent_of_membrane predicted from Blast2Go analysis.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp9-v4.xlsx
-
Supplementary file 10
Relationships among Ks in peak abundance, relative abundance of genes in different ranges of selection pressures (Ka/Ks ratios), and divergence time.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp10-v4.docx
-
Supplementary file 11
Source of leaf materials used for the study.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp11-v4.xlsx
-
Supplementary file 12
Number of genes with different category value of Ks and Ka/Ks in each species pair.
- https://cdn.elifesciences.org/articles/45199/elife-45199-supp12-v4.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/45199/elife-45199-transrepform-v4.docx