Genetic Engineering: Increasing the uptake of carbon dioxide
Look around: how many things do you see made of wood, cloth or plastic? These items may seem wildly different, but they all contain organic carbon and, therefore, they can only exist because plants, algae and certain bacteria are constantly using photosynthesis to turn sunlight, water and atmospheric carbon dioxide (CO2) into most of our food, furniture and fuel (Fischer et al., 2016). However, this process has gotten more difficult over time. Modern CO2 levels are less than 1% of what they were when photosynthetic organisms first evolved, making the work of Rubisco, the enzyme that converts CO2 into organic molecules, more difficult. In turn, the slow rate of CO2 uptake limits the growth of many plants, including crops such as rice and wheat (Long et al., 2006).
Some organisms, however, have evolved ways to concentrate CO2 around Rubisco, allowing the enzyme to run faster (Hennacy and Jonikas, 2020). Introducing such carbon-concentrating mechanisms into crops could increase yields by 60% while reducing water and fertilizer requirements (McGrath and Long, 2014). The best understood carbon-concentrating mechanism is the one found in bacteria, which is based on a protein structure called the ‘carboxysome’ that contains Rubisco and other carbon fixation-related enzymes. These species actively import carbon in the form of bicarbonate (HCO3–), which diffuses into the carboxysome and is converted to CO2. The resulting high CO2 concentration achieved within the carboxysome maximizes the activity of Rubisco and therefore increases overall CO2 uptake (Figure 1A).
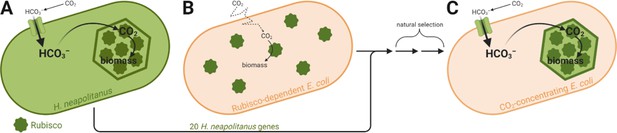
Engineering a carbon-concentrating mechanism into E. coli.
(A) Halothiobacillus neapolitanus has a carbon-concentrating mechanism that relies on structures called carboxysomes. The cell imports CO2 as bicarbonate (HCO3–), which diffuses into the carboxysome (green hexagon) and is converted into concentrated CO2. The elevated levels of CO2 in the carboxysome allow the enzyme Rubisco (dark green) to convert it to biomass more efficiently. (B) Flamholz et al. engineered a strain of E. coli to be dependent on Rubisco activity for its growth. Rubisco runs slowly in this strain as it can only use CO2 which diffuses (dotted line) into the cell from the atmosphere. (C) However, adding just 20 genes from H. neapolitanus and selecting for cells that can grow in low levels of CO2 led to an E. coli strain with a reconstituted H. neapolitanus carbon-concentrating mechanism based on carboxysomes, which allows Rubisco to run much faster.
Figure created with BioRender.com.
Previous work managed to assemble carboxysome-like structures in the non-photosynthetic model bacterium Escherichia coli (Bonacci et al., 2012). However, these cells required high levels of CO2 for growth, indicating that additional components were required to concentrate CO2. Now, in eLife, David Savage, Ron Milo and colleagues – including Avi Flamholz as first author – report how they have engineered a functional carbon-concentrating mechanism into an organism that lacks one (Flamholz et al., 2020).
The team, which is based at the University of California, Berkeley, the Weizmann Institute of Science and the Max Planck Institute of Molecular Plant Physiology, chose the bacterium Halothiobacillus neapolitanus as the genetic donor for their experiment. Carboxysomes in this species are simple and well-studied: in particular, Savage and co-workers had previously identified 20 candidate genes likely needed for these structures to work properly (Desmarais et al., 2019).
As their recipient species, Flamholz et al. chose E. coli, which they genetically modified to rely on Rubisco’s activity for growth (Figure 1B). Without a carbon-concentrating mechanism, this strain could not grow in ambient air – it required supplementation with CO2 levels about 100 times higher than those found in the atmosphere. Hoping to reconstitute a functional carbon-concentrating mechanism, the team transferred the 20 candidate genes from H. neapolitanus to their E. coli strain. Unsurprisingly, the strain was still unable to grow in ambient CO2 at first, as simply adding genes is often not enough to engineer a complex pathway into a new organism (Antonovsky et al., 2016).
However, Flamholz et al. were able to leverage an important feature of their genetically engineered E. coli strain – its growth rate is proportional to Rubisco’s activity. This allowed the team to use a natural selection experiment to spot mutations that make the carbon-concentrating mechanism work, and therefore increase Rubisco activity. The experiment revealed a mutant that could grow at ambient CO2 levels, apparently by adjusting the expression levels of the proteins taking part in the carbon-concentrating process.
This result suggested that a carbon-concentrating mechanism based on H. neapolitanus carboxysomes had successfully been reconstituted in their E. coli strain (Figure 1C). To further support this conclusion, electron microscopy was used to observe the carboxysome-like structures within the engineered E. coli strain. To make sure these structures were functional, they individually knocked out several genes known to be essential for carboxysome function in the native host. These mutations had the same effect in E. coli as in H. neapolitanus – the cells no longer grew at ambient CO2 levels – confirming that the carboxysome was working the same way in the engineered strain as in the native host.
These results from Flamholz et al. indicate that a carboxysome-based carbon-concentrating mechanism can be transferred and function in another organism, providing a blueprint that paves the way toward engineering plants with increased CO2 uptake and thus greater yields.
References
-
DABs are inorganic carbon pumps found throughout prokaryotic phylaNature Microbiology 4:2204–2215.https://doi.org/10.1038/s41564-019-0520-8
-
Evolution of oxygenic photosynthesisAnnual Review of Earth and Planetary Sciences 44:647–683.https://doi.org/10.1146/annurev-earth-060313-054810
-
Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yieldsAnnual Review of Plant Biology 71:461–485.https://doi.org/10.1146/annurev-arplant-081519-040100
-
Can improvement in photosynthesis increase crop yields?Plant, Cell and Environment 29:315–330.https://doi.org/10.1111/j.1365-3040.2005.01493.x
Article and author information
Author details
Publication history
Copyright
© 2020, Franklin and Jonikas
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,750
- views
-
- 204
- downloads
-
- 3
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 3
- citations for umbrella DOI https://doi.org/10.7554/eLife.64380






