Mir155 regulates osteogenesis and bone mass phenotype via targeting S1pr1 gene
Abstract
MicroRNA-155 (miR155) is overexpressed in various inflammatory diseases and cancer, in which bone resorption and osteolysis are frequently observed. However, the role of miR155 on osteogenesis and bone mass phenotype is still unknown. Here, we report a low bone mass phenotype in the long bone of Mir155-Tg mice compared with wild-type mice. In contrast, Mir155-KO mice showed a high bone mass phenotype and protective effect against inflammation-induced bone loss. Mir155-KO mice showed robust bone regeneration in the ectopic and orthotopic model, but Mir155-Tg mice showed compromised bone regeneration compared with the wild-type mice. Similarly, the osteogenic differentiation potential of bone marrow stromal stem cells (BMSCs) from Mir155-KO mice was robust and Mir155-Tg was compromised compared with that of wild-type mice. Moreover, Mir155 knockdown in BMSCs from wild-type mice showed higher osteogenic differentiation potential, supporting the results from Mir155-KO mice. TargetScan analysis predicted sphingosine 1-phosphate receptor-1 (S1pr1) as a target gene of Mir155, which was further confirmed by luciferase assay and Mir155 knockdown. S1pr1 overexpression in BMSCs robustly promoted osteogenic differentiation without affecting cell viability and proliferation. Furthermore, osteoclastogenic differentiation of Mir155-Tg bone marrow-derived macrophages was inhibited compared with that of wild-type mice. Thus, Mir155 showed a catabolic effect on osteogenesis and bone mass phenotype via interaction with the S1pr1 gene, suggesting inhibition of Mir155 as a potential strategy for bone regeneration and bone defect healing.
Editor's evaluation
The authors have shown the important role of miR155 in promoting bone regeneration and higher bone mass. Their fundamental work shows compelling evidence that miR155 is overexpressed in inflammatory diseases therefore, the use of anti-miR155 could produce anti-inflammatory effects. This shows the importance of miR155 inhibitors as therapeutics to promote bone regeneration in inflammatory conditions.
https://doi.org/10.7554/eLife.77742.sa0Introduction
MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs with 18–22 nucleotides length that bind to the 3′-untranslated region of the target gene and regulate the target gene expression (Gareev et al., 2020). miRNAs regulate cell functions such as growth, differentiation, and energy metabolism by silencing the target gene via degradation or translational repression (Gareev et al., 2020; Wang et al., 2019b). Moreover, miRNAs are also involved in the pathophysiology of various inflammatory diseases and cancers (Kumar et al., 2017; Acunzo et al., 2015; Gao et al., 2020). Certain miRNAs had been reported to regulate osteogenesis and bone homeostasis (Gao et al., 2020; Wang et al., 2019a). MicroRNA-155 (miR155) is one of the best conserved and multifunctional miRNAs that regulate several biological processes and diseases such as tumorigenesis, cardiovascular disease, kidney diseases, etc. (Elton et al., 2013; Readhead et al., 2020; Bala et al., 2016; Wu et al., 2018). miR155 is upregulated in inflammatory diseases and cancers, including periodontitis, lung cancer, liver cancer, and breast cancer (Wu et al., 2021; Shao et al., 2019; Xin et al., 2020; Pasculli et al., 2020). Systemic bone loss is frequently observed in patients with inflammatory diseases and cancers (Dimitroulas et al., 2013; Badri et al., 2019). However, the role of miR155 on osteogenesis and bone homeostasis is still unclear.
Induced osteoclasts formation/activity and compromised osteogenic differentiation disrupt bone homeostasis causing bone loss (Kitaura et al., 2020; Li et al., 2014). Osteoclast formation and activity are induced during inflammation and cancer (Adamopoulos, 2018; Roodman, 2001). Mir155 had been reported to induce osteoclastogenesis (Kagiya and Nakamura, 2013). Mir155 knockout (Mir155-KO) mice exhibit reduced local bone destruction in arthritis attributed to reduced generation of osteoclasts (Blüml et al., 2011). Osteogenic differentiation of precursor cells results in bone formation and is the key anabolic event of bone homeostasis. Reduced osteogenic differentiation of precursor cells causes low bone mass phenotype increasing the risk of fracture. Osteogenesis is also a key biological process of bone tissue engineering. Various miRNAs targeted approaches have been developed to promote bone regeneration and bone defect healing during bone tissue engineering (Arriaga et al., 2019). The role of Mir155 in osteogenic differentiation and bone regeneration has been rarely investigated. Compromised osteogenesis and low bone mass phenotype are frequently observed in patients with inflammatory diseases and cancers (Schmidt et al., 2019; Mann et al., 2009). Similarly, effective bone regeneration and bone defect healing are also key challenges in patients with inflammatory diseases. Mir155 targets multiple genes to regulate the pathophysiology of a specific disease in a cell type-specific manner (Hsin et al., 2018). Sphingosine 1-phosphate receptor-1 (S1pr1) is one of the target genes of Mir155 (Xin et al., 2015), which has been reported to positively regulate the osteogenic differentiation of precursor cells (Sato et al., 2012; Higashi et al., 2016). Therefore, it is wise to explore the involvement of S1pr1 in the Mir155-mediated effect on osteogenesis.
In this study, we aimed to analyze the effect of different levels of Mir155 on osteogenesis and bone mass phenotype using Mir155 transgenic (Mir155-Tg) and Mir155-KO mice. This study also investigated the role of Mir155 target gene S1pr1 on the osteogenic differentiation of bone marrow stromal stem cells (BMSCs). We found a catabolic effect of Mir155 on osteogenesis and bone mass phenotype by targeting the S1pr1 gene.
Results
Mir155-Tg mice showed a low bone mass phenotype
We analyzed the expression pattern of Mir155-Tg mice. Mir155 expression was 8.57-fold higher in bone tissue of Mir155-Tg mice compared with the wild-type mice (Figure 1A). Hematoxylin and eosin (H&E) staining shows the growth plate, trabecular bone, cortical bone, and marrow structure of Mir155-Tg and wild-type mice. Low trabecular density was observed in Mir155-Tg mice compared with wild-type mice (Figure 1B). Micro-CT results showed reduced cortical bone thickness and bone volume/total volume (BV/TV) in Mir155-Tg mice (Figure 1C and D). The bone mineral density (BMD) level was not significantly changed (Figure 1D). Micro-CT images showed fewer and thinner trabeculae in Mir155-Tg mice femur compared with wild-type mice (Figure 1E). Trabecular bone parameter BV/TV, BMD, trabecular number (Tb.N), and trabecular thickness (Tb.Th) were significantly reduced in Mir155-Tg mice compared with wild-type mice (Figure 1F). In Mir155-Tg mice, trabecular separation (Tb.Sp) was significantly increased compared with wild-type mice (Figure 1F). These results indicate the low bone mass phenotype in Mir155-Tg mice.
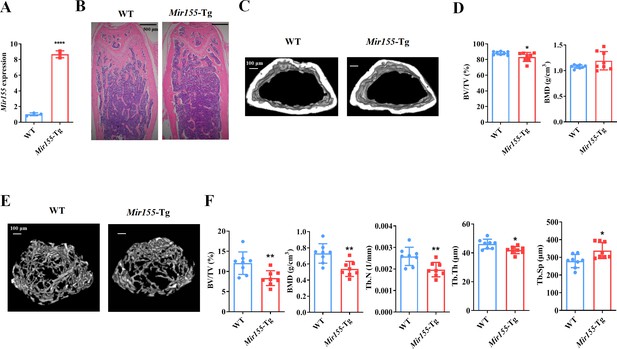
Mir155 transgenic (Mir155-Tg) mice showed a low bone mass phenotype.
(A) Mir155 expression in bone, (B) Hematoxylin and eosin (H&E) staining, (C) representative micro-CT images for cortical bone, (D) bone volume/total volume (BV/TV) and bone mineral density (BMD) analysis, (E) representative micro-CT images for trabecular bone, (F) BV/TV, BMD, trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp) analysis. Data are presented as mean ± SD, n=8. Significant difference compared to wild-type mice, *p<0.05, **p<0.01, and ****p<0.0001.
-
Figure 1—source data 1
Raw data for Figure 1A, D (BV/TV and BMD), and Figure 1F (BV/TV, BMD, Tb.N, Tb.Th, and Tb.Sp).
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig1-data1-v2.xlsx
Mir155-KO mice showed a high bone mass phenotype
Mir155 expression in Mir155-KO mice was dramatically downregulated compared with wild-type mice (Figure 2A). H&E staining demonstrated that trabeculae were increased in the Mir155-KO mice compared with wild-type mice (Figure 2B). Micro-CT results showed a higher cortical bone thickness and BV/TV in Mir155-KO mice (Figure 2C and D). The BMD level was not significantly increased in Mir155-KO mice compared with wild-type (Figure 2D). Micro-CT images showed robustly dense and interconnected trabeculae in Mir155-KO mice compared with wild-type mice (Figure 2E). The trabecular bone parameters BV/TV, BMD, and Tb.N in Mir155-KO mice were increased by 2-, 2.69-, and 1.83-fold respectively, compared with wild-type mice (Figure 2F). While Tb.Th was similar in Mir155-KO and wild-type mice (Figure 2F). Tb.Sp in Mir155-KO mice was significantly reduced compared with wild-type mice (Figure 2F). These results indicate the high bone mass phenotype of Mir155-KO mice. Mir155-KO and Mir155-Tg showed an opposite trend of bone mass phenotype and bone parameters (Figures 1 and 2), suggesting the role of Mir155 in bone homeostasis regulation.
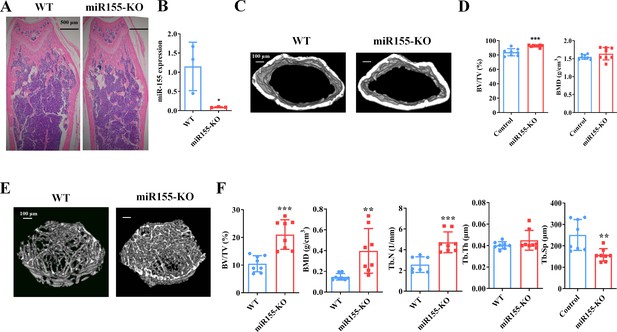
Mir155 knockout (Mir155-KO) mice showed a high bone mass phenotype.
(A) Mir155 expression in bone, (B) Hematoxylin and eosin (H&E) staining, (C) representative micro-CT images for cortical bone, (D) bone volume/total volume (BV/TV) and bone mineral density (BMD) analysis, (E) representative micro-CT images for trabecular bone, (F) BV/TV, BMD, trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp) analysis. Data are presented as mean ± SD, n=8. Significant difference compared to wild-type group, *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 2—source data 1
Raw data for Figure 2A, D (BV/TV and BMD), and Figure 2F (BV/TV, BMD, Tb.N, Tb.Th, and Tb.Sp).
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig2-data1-v2.xlsx
Lipopolysaccharide-related osteolysis
We analyzed the effect of Mir155-KO in the context of inflammation-related bone loss. H&E staining of long bone tissue sections showed more trabecular bone in lipopolysaccharide (LPS)-treated Mir155-KO mice compared with LPS-treated wild-type mice (Figure 3A). Micro-CT images showed slightly thicker cortical bone in LPS-treated Mir155-KO mice compared with LPS-treated wild-type mice (Figure 3B). Cortical bone parameters BV/TV and BMD were significantly increased in LPS-treated Mir155-KO mice compared with LPS-treated wild-type mice (Figure 3C). Micro-CT images showed more dense and interconnected trabeculae in LPS-treated Mir155-KO mice compared with LPS-treated wild-type mice (Figure 3D). The trabecular bone parameters BV/TV, BMD, and Tb.N were increased by 2.3-, 1.22-, and 2.68-fold, respectively, in LPS-treated Mir155-KO mice compared with LPS-treated wild-type mice (Figure 3E). Tb.Sp was significantly decreased in LPS-treated Mir155-KO mice compared with LPS-treated wild-type mice (Figure 3E). These results demonstrate the protective effect of Mir155-KO against LPS-induced bone loss.
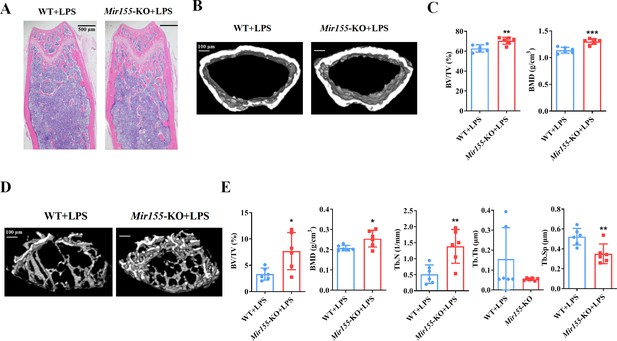
Mir155 knockout (Mir155-KO) mice showed higher resistance against lipopolysaccharide (LPS)-induced bone loss.
(A) Hematoxylin and eosin (H&E) staining, (B) representative micro-CT images for cortical bone, (C) bone volume/total volume (BV/TV) and bone mineral density (BMD) analysis, (D) Representative micro-CT images for trabecular bone, (E) BV/TV, BMD, trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp) analysis. Data are presented as mean ± SD, n=6. Significant difference compared to wild-type group, *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 3—source data 1
Raw data for Figure 3C (BV/TV and BMD) and Figure 3E (BV/TV, BMD, Tb.N, Tb.Th, and Tb.Sp).
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig3-data1-v2.xlsx
Ectopic bone regeneration was inhibited in Mir155-Tg mice while increased in Mir155-KO mice
A bone regeneration study was conducted to investigate whether the bone regeneration potential is altered in Mir155-Tg and Mir155-KO mice. BMP2-loaded collagen membranes were implanted in mice ectopically to confirm the bone regeneration potential in the ectopic site of Mir155-Tg, Mir155-KO, and the respective wild-type mice. Micro-CT images showed very less bone volume in collagen membrane transplanted in Mir155-Tg mice compared with that of wild-type mice (Figure 4A). The Mir155-Tg group showed significantly reduced BV/TV and Tb.N in newly formed bone compared with the wild-type group (Figure 4B and C). Tb.Th and Tb.Sp levels were similar in the Mir155-Tg and wild-type groups (Figure 4D and E). In contrast, ectopic bone regeneration was significantly increased in the Mir155-KO group compared with the wild-type group (Figure 4F). Newly formed bone BV/TV and Tb.N in the Mir155-KO group were increased by 6.12- and 5.64-fold respectively compared with the wild-type group (Figure 4G and H). Tb.Th remained unchanged in Mir155-KO mice (Figure 4I) but the Tb.Sp was significantly reduced in the Mir155-KO group compared with the wild-type group (Figure 4J). These results indicate a catabolic effect of Mir155 on bone regeneration.
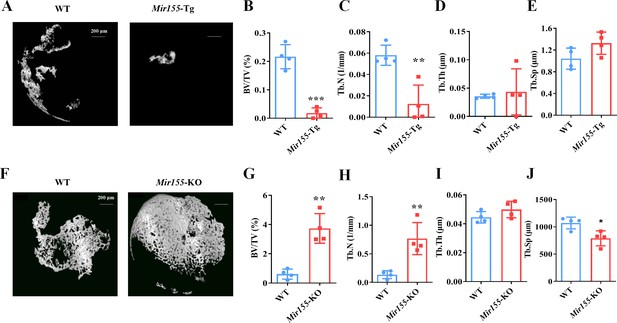
Ectopic bone regeneration was inhibited in Mir155 transgenic (Mir155-Tg) mice but enhanced in Mir155 knockout (Mir155-KO) mice.
(A) Representative micro-CT images. (B) Bone volume/total volume (BV/TV), (C) trabecular number (Tb.N), (D) trabecular thickness (Tb.Th), and (E) trabecular separation (Tb.Sp) analysis in Mir155-Tg and wild-type mice. (F) Representative micro-CT images. (G) BV/TV, (H) Tb.N, (I) Tb.Th, and (J) Tb.Sp analysis in Mir155-KO and wild-type mice. Data are presented as mean ± SD, n=4. Significant difference compared to wild-type group, *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 4—source data 1
Raw data for Figure 4B, E, G-J.
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig4-data1-v2.xlsx
Mir155-KO mice showed enhanced bone regeneration in an orthotopic model
We further added a low dose of BMP-2 to the collagen membrane and evaluated the bone regeneration in calvarial bone defect of Mir155-KO and wild-type mice. The defect area was covered with a robustly high amount of newly formed bone in Mir155-KO mice compared with wild-type mice (Figure 5A and B). Furthermore, BV/TV, BMD, and Tb.N (Figure 5C–E) were enhanced while Tb.Th and Tb.Sp were reduced in Mir155-KO mice compared with wild-type mice (Figure 5F and G). These results from calvarial bone defect healing analysis showed promising bone regeneration effects of Mir155-KO.
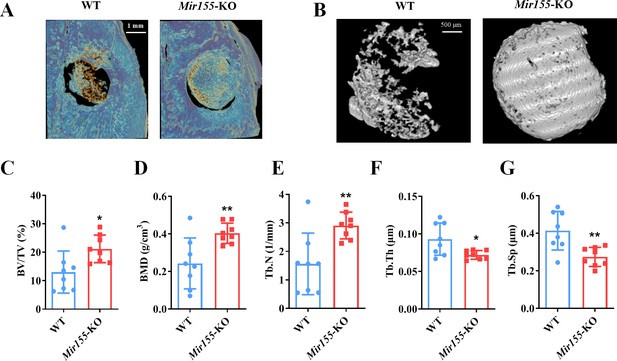
A higher degree of bone regeneration was observed in the calvarial defect of Mir155 knockout (Mir155-KO) mice with a low dose of BMP2 treatment.
(A) Representative micro-CT images, (B) local micro-CT images in defects, (C) bone volume/total volume (BV/TV), (D) bone mineral density (BMD), (E) trabecular number (Tb.N), (F) trabecular thickness (Tb.Th), and (G) trabecular separation (Tb.Sp) analysis. Data are presented as mean ± SD, n=8. Significant difference compared to wild-type mice, *p<0.05 and **p<0.01.
-
Figure 5—source data 1
Raw data for Figure 5C-G.
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig5-data1-v2.xlsx
Mir155 influences the osteogenic differentiation of BMSCs
To further confirm the regulatory role of Mir155 on osteogenesis, we analyzed the osteogenic differentiation potential BMSCs isolated from Mir155-Tg, Mir155-KO, and the respective wild-type mice. Mineralized matrix deposition potential in BMSCs from Mir155-TG mice was substantially reduced compared with that of wild-type mice (Figure 6A and B). Similarly, protein expression levels of osteogenic markers ALP and RUNX2 in BMSCs from Mir155-Tg mice were reduced compared with those of wild-type mice (Figure 6C and D). These results indicate the compromised osteogenic differentiation potential of BMSCs from Mir155-Tg mice. In contrast, BMSCs from Mir155-KO mice showed robustly higher matrix mineralization potential compared to those of wild-type mice (Figure 6E and F). The protein expression levels of osteogenic markers ALP were enhanced in BMSCs from Mir155-KO (Figure 6G and H). However, RUNX2 protein levels were not changed in BMSCs from Mir155-KO mice compared with those from wild-type mice. These results demonstrated the catabolic effect of Mir155 in the osteogenic differentiation of BMSCs.
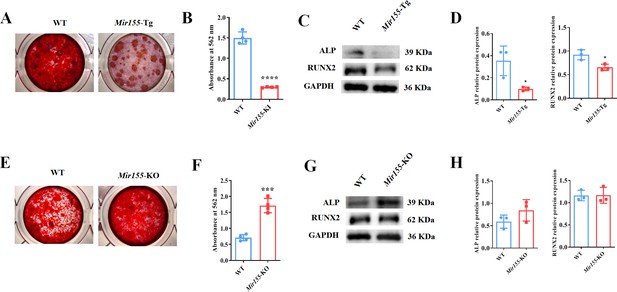
Mir155 transgenic (Mir155-Tg) and Mir155 knockout (Mir155-KO) bone marrow stromal stem cells (BMSCs) showed an opposite trend of osteogenic differentiation.
(A) Alizarin red staining (ARS) images at day 10 of culture, (B) ARS quantification, n=4, (C) Western blot analysis of osteogenic markers, and (D) densitometry quantification of protein bands in Mir155-Tg BMSCs, n=3. (E) ARS images stained at day 10 of culture, (F) ARS quantification, n=4, (G) Western blot analysis of osteogenic markers, and (H) densitometry quantification of protein bands in Mir155-KO BMSCs, n=4. Data are presented as mean ± SD. Significant difference compared to wild-type mice, *p<0.05, ***p<0.001, and ****p<0.0001.
-
Figure 6—source data 1
Raw data for Figure 6B, D (ALP and RUNX2), Figure 6E, H (ALP and RUNX2); original blots for Figure 6C and G.
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig6-data1-v2.zip
Mir155 knockdown promotes the osteogenic differentiation of BMSCs
Mir155 knockdown in BMSCs was used to further confirm the role of Mir155 in osteogenic differentiation. Mir155 sponge lentivirus treatment significantly reduced the expression of Mir155 in BMSCs (Figure 7A), indicating the successful knockdown of Mir155. Matrix mineralization was robustly increased in the Mir155 sponge group compared with the negative control group (Figure 7B and C). Furthermore, similarly, the protein expression levels of osteogenic markers ALP and RUNX2 were robustly upregulated in the Mir155 sponge group compared with the negative control group (Figure 7D and E). Furthermore, sponging Mir155 did not affect cell viability and expression of mesenchymal stem cell markers in BMSCs (Figure 7F and G). These results further confirm the catabolic effect of Mir155 on the osteogenic differentiation of precursor cells.
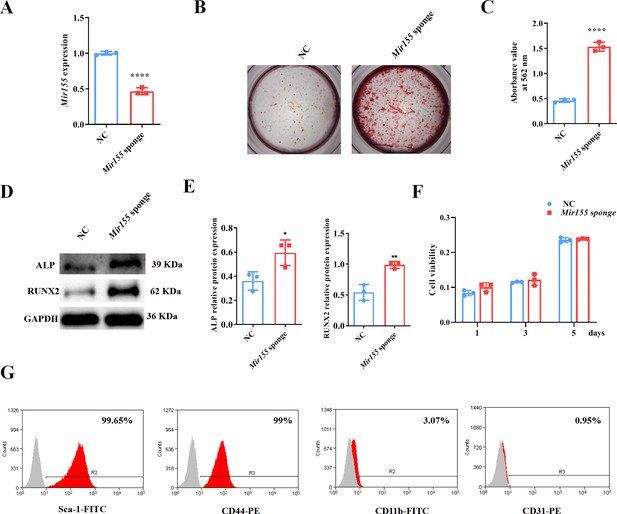
Mir155 knockdown bone marrow stromal stem cells (BMSCs) showed higher osteogenic differentiation potential.
(A) Mir155 expression level, n=3, (B) Alizarin red staining (ARS) images stained at 21 days of culture, (C) ARS quantification, n=3, (D) Western blot analysis, (E) densitometry quantification of protein bands, n=3, (F) cell viability, n=3, and (G) Fluorescence activated cell sorting (FACS) analysis. Data are presented as mean ± SD. Significant difference compared to the negative control, *p<0.05, **p<0.01, and ****p<0.0001. NC: negative control.
-
Figure 7—source data 1
Raw data for Figure 7A, C, E (ALP and RUNX2), and Figure 7F; original blots for Figure 7D.
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig7-data1-v2.zip
Mir155 targets the S1pr1 gene to regulate the osteogenic differentiation of BMSCs
TargetScan prediction showed that the binding sites of Mir155 on S1pr1 were rather conserved in different species, such as human, mice, rat, rhesus, etc. (Figure 8A). The sequences of seeding sites and mutant seeding sites of S1pr1 are shown in Figure 8B. Luciferase reporter gene assay was performed to analyze the Mir155 and S1pr1 gene interaction (Figure 8C). Our results showed that Mir155 directly binds to the 3’UTR of the S1PR1 (Figure 8C). Sponging Mir155 robustly enhanced the protein level expression of the S1pr1 gene in BMSCs (Figure 8D), confirming the interaction of Mir155 and the S1pr1 gene. S1pr1 transfection in BMSCs robustly enhanced the matrix mineralization (Figure 8E and F). The protein expression level of S1PR1 was enhanced in lentivirus-mediated S1pr1 overexpressed BMSCs (Figure 8G and H). This result indicates the efficacy of lentivirus-based S1pr1 overexpression in BMSCs. The protein expression levels of ALP and RUNX2 were increased in S1pr1 overexpressed BMSCs (Figure 8G and H). S1pr1 overexpression in BMSCs by lentivirus did not affect cell viability and proliferation (Figure 8I). These results indicate that the Mir155 targets the S1pr1 gene to regulate the osteogenic differentiation of BMSCs.
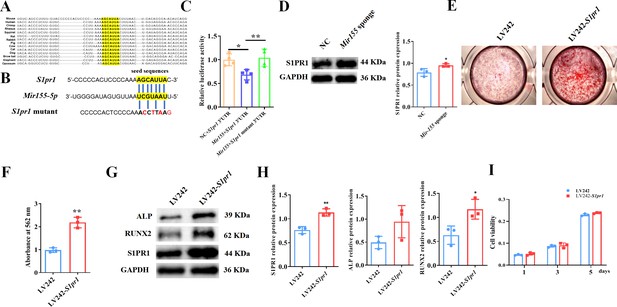
Mir155 targets sphingosine 1-phosphate receptor-1 (S1PR1) to regulate the osteogenic differentiation of bone marrow stromal stem cells (BMSCs).
(A) The Mir155 binding site of S1pr1 in different species, (B) the wild and mutant binding site of S1pr1 in mice. (C) Luciferase assay, n=4, (D) S1PR1 protein expression and densitometry quantification of protein bands, n=3, (E) Alizarin red staining (ARS) images stained at 21 days of culture, (F) ARS quantification, n=3, (G) Western blot analysis of osteogenic markers, (H) densitometry quantification of protein bands, n=3, and (I) cell viability analysis, n=4. Data are presented as mean ± SD. Significant difference compared to negative control, *p<0.05 and **p<0.01.
-
Figure 8—source data 1
Raw data for Figure 8C, D, F, H and I; original blots for Figure 8D, G.
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig8-data1-v2.zip
Mir155 influences osteoclastogenesis
The function of Mir155 on osteoclastogenesis was further explored. Primary bone marrow monocytes were isolated and induced into bone marrow-derived macrophages (BMMs). The receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL) was further used for osteoclastogenic differentiation. Tartrate-resistant acid phosphatase (TRAP) staining showed that Mir155-KO reduced the osteoclast number (Figure 9A and B). Gene expression of osteoclastogenesis markers including receptor activator of NF-κB (Rank) and cathepsin K (Ctsk) was significantly reduced during the osteoclastogenic differentiation of Mir155-KO BMMs (Figure 9C). Furthermore, bone resorption-related C-terminal telopeptides of type I collagen (CTX-1) level were significantly reduced while bone formation-related procollagen type I N-terminal pro-peptide (PINP) level was increased in serum of Mir155-KO mice compared with wild-type mice (Figure 9D). These results demonstrate that Mir155-KO has the potential inhibitory effect on osteoclastogenesis.
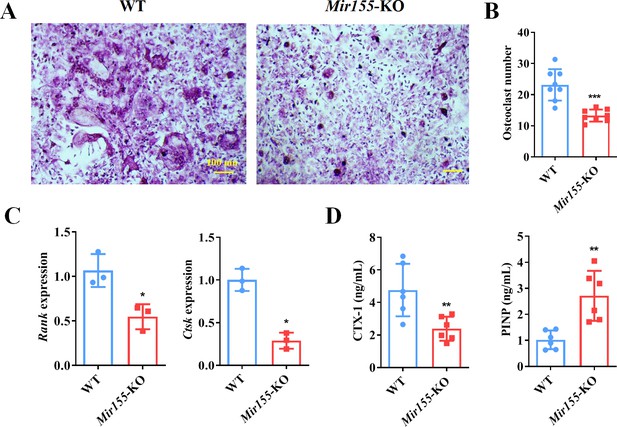
Bone marrow-derived macrophages (BMMs) from Mir155 knockout (Mir155-KO) mice exhibited compromised osteoclastogenic differentiation.
(A) Tartrate-resistant acid phosphatase (TRAP) staining, (B) osteoclast number quantification, n=8, (C) osteoclastogenic markers expression, n=3, (D) C-terminal telopeptides of type I collagen (CTX-1), and procollagen type I N-terminal pro-peptide (PINP) serum level, n=6. Data are presented as mean ± SD. Significant difference compared to the negative control, *p<0.05, **p<0.01, and ***p<0.001.
-
Figure 9—source data 1
Raw data for Figure 9B, C and D.
- https://cdn.elifesciences.org/articles/77742/elife-77742-fig9-data1-v2.xlsx
Discussion
Differentiation of MSCs to osteoblasts is a vital event of bone regeneration. Differentiated osteoblasts deposit mineralized matrix and contribute to new bone formation. Various miRNAs have been reported to regulate osteogenesis and bone mass phenotype (Arriaga et al., 2019). In this study, Mir155-Tg mice showed compromised bone regeneration and low bone mass phenotype. In contrast, Mir155-KO mice showed improved bone regeneration, higher bone mass phenotype, and a protective effect against inflammation-induced bone loss. BMSCs from Mir155-Tg and Mir155-KO mice showed compromised and robust osteogenic differentiation potential, respectively. Mir155 knockdown also promoted osteogenic differentiation potential in BMSCs. These results indicate a catabolic effect of Mir155 on bone regeneration and bone mass phenotype. Knockdown of Mir155 in BMSCs robustly enhanced the protein level expression of S1PR1 and osteogenic regulator RUNX2, indicating S1PR1 as a target gene of Mir155 in BMSCs to regulate osteogenic differentiation. S1pr1 overexpression in BMSCs enhanced RUNX2 expression and osteogenic differentiation of BMSCs indicating the regulatory role of Mir155-S1PR1 interaction on osteogenesis (Figure 10).
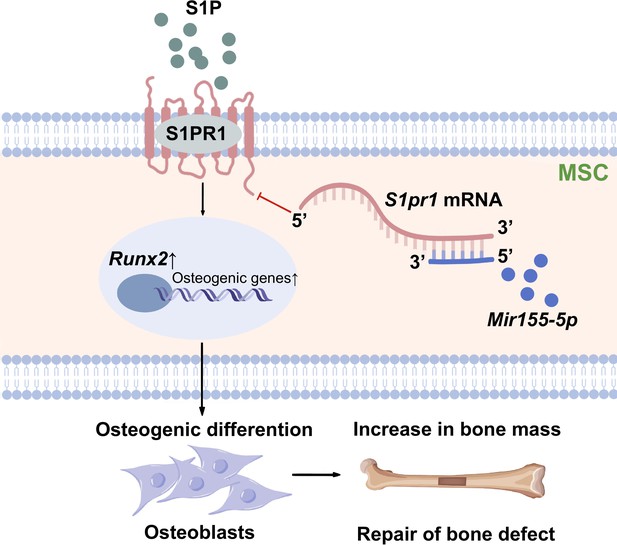
Scheme of Mir155-mediated regulation of osteogenesis.
S1P activates the sphingosine 1-phosphate receptor-1 (S1PR1), further increasing RUNX2 expression to regulate the osteogenic differentiation of MSCs into osteoblasts. Mir155 inhibits this process by direct binding with 3’UTR S1pr1 mRNAs. MSCs: mesenchymal stromal cells.
miRNAs/anti-miRNAs have been used for bone tissue engineering (Arriaga et al., 2019). MiR26a (Wang et al., 2015), anti-miR31 (Deng et al., 2013), anti-miR34a (Chen et al., 2020b), miR135 (Xie et al., 2016), anti-miR138 (Eskildsen et al., 2011), anti-miR146a (Xie et al., 2017), miR148a (Li et al., 2017), anti-miR221 (Yeh et al., 2016), and anti-miR3555p Tomé et al., 2011 have shown an anabolic effect on osteogenic differentiation of precursor cells and bone regeneration. Most of the miRNAs/anti-miRNAs promote bone regeneration via the activation of the osteogenic master regulator RUNX2 (Arriaga et al., 2019). In this study, Mir155 overexpression showed a catabolic effect on osteogenesis and bone mass phenotype. Interestingly, knockout or knockdown of Mir155 showed an anabolic effect on osteogenesis, bone regeneration, and bone defect healing. RUNX2 was involved in the Mir155-mediated regulation of osteogenic differentiation of BMSCs. RUNX2 expression did not change in Mir155-KO BMSCs but was upregulated in Mir155 sponged BMSCs from wild-type mice. Whole body knockout of Mir155 also knock outs the Mir155 in the other cell types present in bone microenvironment, for example, monocytes and macrophages. The Mir155-KO BMSCs might have certain effects of surrounding Mir155-KO other cell types on S1PR1 expression, while the Mir155 sponged BMSCs from wild-type mice sorted with FACS may be free from such effects. This could be the possible explanation for the lack of change in RUNX2 expression in Mir155-KO BMSCs, but rather an upregulation of RUNX2 expression in Mir155 sponged BMSCs from wild-type mice observed in this study. However, further studies are required to support this logic. Mir155 had been reported to inhibit osteogenesis of MC3T3-E1 cells via SMAD5 downregulation (Gu et al., 2017). Mir155 inhibits BMP9-induced osteogenic differentiation of precursor cells via downregulation of BMP signaling (Liu et al., 2018). Qu et al. revealed that miR155 inhibition alleviates high glucose and free fatty acid-suppressed osteogenic differentiation of BMSCs by targeting SIRT1 (Qu et al., 2020). Our results from Mir155-Tg mice on inhibition of osteogenesis are in accordance with the findings from the literature (Gu et al., 2017; Liu et al., 2018). Furthermore, Mir155-KO inhibited the osteoclastogenic differentiation of BMMs. To our knowledge, this is the first study to report the anabolic effect of Mir155-KO or knockdown on osteogenic differentiation and the catabolic effect of Mir155-KO on osteoclastogenesis. Our results suggest the potential application of anti-miR155 on bone regeneration and bone tissue engineering applications.
Anti-miR155 oligonucleotides and antagomir have been designed for various cancer treatments (Kardani et al., 2020; Witten and Slack, 2020). Small molecule-based cyclic peptidomimetics had shown an inhibitory effect on miR155 biogenesis (Yan et al., 2019). MLN4924 is an inhibitor of the NEDD8-activating enzyme. MLN4924 decreases the binding of NF-κB to the miR155 promoter and downregulates miR155 in AML cells (Khalife et al., 2015). Since the knockout of Mir155 promoted bone regeneration and protected against inflammation-induced bone loss, anti-Mir155 or Mir155 inhibitors could be applied for bone tissue engineering and the treatment of low bone mass phenotype and inflammation-related bone loss. However, the osteoinductive potential of already available anti-Mir155 or Mir155 inhibitors should be tested using in vitro and in vivo models to prove this hypothesis.
miR155 targets different genes in different cells to regulate the cell type-specific functions (Woeller et al., 2019; Wang et al., 2018; Hawez et al., 2019; Li et al., 2021c). S1pr1, a target gene of Mir155, is regulated during various physiological and pathological conditions (Li et al., 2021c; Okoye et al., 2014). TargetScan prediction results showed that the binding site of Mir155 on S1pr1 was rather conserved in several different species such as human, rat, mouse, etc. In this study, Mir155 inhibition upregulated S1PR1 protein expression. Overexpression of S1pr1 robustly promoted RUNX2 protein expression and osteogenic differentiation of BMSCs. Mir155 has been shown to inhibit the osteogenic differentiation of precursor cells via inhibiting SMAD5 (Gu et al., 2017). Higashi et al. reported SMAD1/5/8 as downstream signaling of S1PR1/S1PR2 to induce RUNX2 expression in osteoblasts (Higashi et al., 2016). Reports from the literature and results of this study indicate that Mir155 targets the S1pr1 gene to inhibit RUNX2 expression thereby reducing bone regeneration and bone mass.
Since miR155 is upregulated in various cancers including hematological cancers (Witten and Slack, 2020). Hematological cancer mainly affects bone marrow which is the dwelling of bone precursor cells. Hematological cancers are associated with bone loss and fracture of vertebrae and long bones. Breast and lung cancer are frequently metastasized to bone and cause osteolysis (Pang et al., 2020; Cheng et al., 2021). Cancer/cancer metastasis-induced bone loss-mediated fracture is a serious clinical problem. However, the role of upregulated levels of miR155 on cancer/cancer metastasis-related reduced bone mass is still unclear. Moreover, the prevention of cancer/cancer metastasis-induced bone loss is a huge challenge for clinicians. Since anti-miR155 has already been proven to be beneficial for cancer treatment and miR155 knockdown promotes bone regeneration, anti-miR155 could treat cancer as well as caner-induced bone loss as a killing two birds with one stone concept. However, future in vitro and in vivo studies are needed to confirm this hypothesis.
Mir155 is overexpressed and plays a key role in the pathophysiology of inflammatory diseases including autoimmune arthritis, osteoarthritis, and periodontitis (Wu et al., 2021; Blüml et al., 2011; Li et al., 2021a). An elevated level of Mir155 in arthritis promotes M1 macrophage polarization and inflammation (Li et al., 2021a). Prevention of bone loss in inflammatory diseases using currently available therapeutic approaches is not satisfactory. Moreover, inflammation impedes bone regeneration thereby causing the failure of bone tissue engineering approaches. In the present study, we found that Mir155-KO exerts a protective effect against LPS-induced bone loss. Since anti-Mir155 has anti-inflammatory (Teng et al., 2020) and bone regenerative potential, anti-Mir155 could be a potential therapeutic to prevent inflammation-induced bone loss.
This study used both Mir155-Tg and Mir155-KO mice to investigate the role of Mir155 on osteogenesis and bone mass phenotype. Bone regeneration in both ectopic and orthotopic models confirmed the regulatory role of Mir155 in bone regeneration. Mir155 silenced and S1pr1 overexpressed BMSCs further confirmed the S1pr1 as a target gene of Mir155 to regulate RUNX2 expression during osteogenesis. The limitation of this study is that we did not analyze the downstream signaling pathway of S1PR1 that regulates RUNX2 expression in BMSCs. Clinical application of osteogenic factors in vivo always poses the risk of vascular calcification. miR155-5p overexpression has been reported to aggravate vascular calcification (He et al., 2020). Importantly, Mir155-KO mice also show resistance against vitamin D3-induced vascular calcification (Li et al., 2021b). Moreover, Mir155 deletion inhibits the migration and apoptosis of vascular smooth muscle cells as well as vascular calcification (Li et al., 2021b). Furthermore, Zhang et al. showed normal histology of vital organs including the heart, lung, liver, and spleen in Mir155-KO mice (Zhang et al., 2017). These reports from the literature indicate that Mir155-KO does not pose the risk of vascular calcification. However, the effect of Mir155 inhibitors or anti-Mir155 on vascular calcification should be thoroughly investigated before applying these agents for bone regeneration applications.
In conclusion, Mir155 showed a catabolic effect on osteogenesis and bone mass via targeting S1pr1. Our results suggest miR155 as a potential target to promote bone regeneration and higher bone mass. Since miR155 is overexpressed in inflammatory diseases and anti-miR155 has shown anti-inflammatory potential, the miR155 inhibitors could be the potential therapeutics to promote bone regeneration even in inflammatory conditions.
Materials and methods
Mice
Mir155-KO mice were purchased from the Jackson Laboratory (Stock No. 007745). Mir155-Tg mice were constructed as described in our previous reports (Lin et al., 2016). The C57BL/6J wild-type mice, as the wild-type mice of Mir155-KO mice, were purchased from Guangdong Medical Laboratory Animal Center. While the FVB mice were a littermate control of Mir155-Tg mice. The blinded evaluation was used for mice assignments and analysis. The animal experiment was conducted in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China (2017-078).
Bone phenotype analysis
Request a detailed protocolBone phenotype analysis was performed in 8-week-old mice (8 mice/group including 4 male and 4 female mice) using micro-CT. Mice were anesthetized using isoflurane (RWD Life Science Co., China), followed by cervical dislocation. The femur with a distal growth plate was collected and fixed in 10% buffered formalin. Micro-CT scanning was performed to evaluate bone phenotype using Bruker Sky1172 Skyscan (Kontich, Belgium). A total of 100 slices (1 mm) below the distal growth plate of the femurs was measured for 3D reconstruction and quantification of trabecular bone and cortical bone as described previously (Moussa et al., 2021). The X-ray tube was operated at 96 kV and 65 μA using a 0.5 mm Al filter with a resolution of 7.93 μm pixels. Scanning was performed by 180° rotation around the vertical axis, camera exposure time of 1300 ms, rotation step of 0.6°, frame averaging of 2, and random movement of 10. 3D images were made using CTvox software (Skyscan, Kontich, Belgium). Data viewer software (Skyscan, Kontich, Belgium) was used for images and linear analysis. Relative bone formation parameters including BV/TV, BMD, Tb.N, Tb.Th, and Tb.Sp were analyzed.
LPS-treated mice
Request a detailed protocolAs reported by Chen et al., 2020a, wild-type mice and Mir155-KO mice (6 mice/group including 3 male mice and 3 female mice) were injected intraperitoneally with LPS (8 mg/kg) twice 1 week. Mice were weighed before LPS injection. After 6 weeks, femurs were collected for micro-CT analysis and H&E staining.
H&E staining
Request a detailed protocolH&E staining for bone tissues was performed as previously reported (Chen et al., 2020a; Chen et al., 2018). Femurs were fixed in 4% PFA for 2 days and decalcified with EDTA decalcified solution for 21 days. After that, femurs were cut for histological analysis, the femurs were dissected and conducted in 4% PFA for 2 days. Then slices of bone tissue at 3 μm thickness were cut along the coronal plate. The decalcified slices were further performed H&E staining by dewaxing, hematoxylin staining, eosin staining, and dehydration.
Ectopic grafting of collagen membrane
Request a detailed protocolThe subcutaneous transplantation of the collagen membrane was performed as described previously (Huang et al., 2017). Collagen membrane ZH-BIO (China) with 5 mm diameter and 1 mm thickness were osteogenically functionalized by loading 10 μL of 0.3 mg/mL BMP2 solution. BMP2-loaded collagen membranes were implanted in the subcutaneous pockets of Mir155-Tg, Mir155-KO, and respective wild-type mice. Eight-week-old male mice with 20–22 g body weight (4 mice/group, 1 membrane/mouse) were used for this study. After 18 days of transplantation, mice were euthanized by isoflurane, collagen membrane was collected and further analyzed for newly formed bone using micro-CT.
Mice calvaria bone defect healing with BMP2 addition
Request a detailed protocolAs previously reported (Reyes et al., 2018), a 3 mm diameter was generated on one side of the sagittal suture in 8-week Mir155-KO mice and wild-type mice (8 mice/group including 4 male mice and 4 female mice, 1 defect/mouse). A total of 200 ng BMP2 (10 μL) was dropped into the collagen membrane ZH-BIO (China). The membrane was inserted into the defect. Calvaria bones were collected and analyzed by micro-CT after 1 month.
The isolation of primary BMSCs and osteoclastogenic induction
Request a detailed protocolEuthanized transgenic mice and wild-type male mice (5–6 weeks of age) were immersed into 75% ethanol for 5 min. The femurs and tibia were acquired. Primary BMSCs were isolated and expanded as described previously (Soleimani and Nadri, 2009). In brief, bone marrow was flushed out from the tibia and femurs and disturbed into small pieces. Cells were collected by centrifugation and plated into flasks and allowed to adhere for 24 hr. Nonadherent cells were washed, and culture was continued in DMEM supplemented with 10% non-heat inactivated FBS and 1% penicillin/streptomycin. The cells were cultured in a 5% CO2 incubator maintaining a humid atmosphere. Cells were trypsinized from 80% confluent culture and passaged.
Analysis of the target gene of Mir155
Request a detailed protocolTargetScan software was used to predict the target gene of Mir155. TargetScan predicted S1PR1 as a possible target gene of Mir155.
Plasmid construction and lentivirus preparation
Request a detailed protocolThe S1pr1 3’UTR sequences and mutant sequences (200 bp upstream and 200 bp downstream of the binding site from NM_007901.5 transcript) were synthesized and cloned into wild-type plasmid pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, USA) by Generay (China). We created S1pr1 3’UTR and S1pr1 mutant 3’UTR plasmids for luciferase assay. The LV242 S1pr1 and control LV242 plasmid were purchased from Genecopedia (USA). Mir155 negative control (NC) and Mir155 sponge plasmids were purchased from OBIO (China).
As previously described (Zheng et al., 2013), HEK293T cells were co-transfected with expression plasmids (Mir155 NC, Mir155 sponge, LV242, or LV242 S1pr1) with the packaging plasmids pMD2.VSVG, pMDLg/pRRE, and pRSV-REV using EZ trans transfection regent (Shanghai life iLab BioTechnology Co., Ltd., China). After 48 hr, fresh lentiviral supernatant was collected and used for infection. BMSCs were expanded to 60% confluence before lentiviral infection. After infection for 10 hr, cells were washed and allowed to recover for 24 hr and used for subsequent experiments. Mir155 sponge efficacy was analyzed by RT-qPCR. S1PR1 overexpression efficacy was analyzed by Western blot analysis.
Luciferase assay
Request a detailed protocolLuciferase assay was performed as previously described (Wang et al., 2019c). Luciferase assay was performed for further confirmation of S1pr1 as a target gene of Mir155. S1PR1 3’UTR (100 ng) with NC (50 nM), S1pr1 3’UTR (100 ng) with Mir155 (50 nM), and the S1pr1 mutant 3’UTR plasmid (100 ng) with Mir155 (50 nM) were co-transfected into HEK293T cells by Lipofectamine 2000 (Thermo Fisher Scientific Inc, USA). After 48 hr, the luciferase assay was performed according to the manufacturer’s instructions using Luc-Pair Duo-Luciferase Assay Kit 2.0 (GeneCopoeia, USA). In brief, the cells were lysed and incubated with 100 μL Fluc work solution for 5 min, the fluorometric measurement was performed by Varioskan Flash (Thermo Fisher, USA). The lysis solution was added with 100 μL Rluc, incubated for 5 min, and the fluorometric value was measured.
Alizarin red staining
Request a detailed protocolAlizarin red staining was performed as a previous report (Wang et al., 2022). BMSCs (28,000 cells/well) were seeded at 48-well culture plates and cultured with osteogenic medium (50 μg/mL vitamin C, 0.01 μM dexamethasone, and 10 mM β-glycerophosphate). Then, the cells were fixed with paraformaldehyde and stained with Alizarin Red S solution (1%, pH 4.2) (Solarbio Life Science, China) for 10 min. The staining was visualized under stereomicroscope Leica EZ4HD (Leica, Germany). For quantitative analysis, the alizarin red-stained mineralized matrix was dissolved with 200 μL 10% hexadecylpyridinium chloride monohydrate for 1 hr, and the supernatant was collected. The optical density of the supernatant (100 μL) was measured by a microplate reader at 562 nm.
Immunoblotting
Request a detailed protocolImmunoblotting was performed as a previous report (Zhang et al., 2021). Cells were lysed by using RIPA Buffer (CWBio, China) containing protease inhibitors to extract total protein. Total protein (20 μg) was added to 10% SDS-polyacrylamide gel. The protein was transferred to PVDF membranes (Millipore, USA) after electrophoresis and blocked for 1 hr with a blocking buffer (Beyotime, China). Then PVDF membranes were incubated with the primary antibodies including ALP (Abcam, UK, 1:3000), RUNX2 (CST, USA, 1:2,000), S1PR1 (Abcam, UK, 1:2,000), and GAPDH (CST, USA, 1:5,000) overnight at 4°C. The membranes were further incubated with horseradish peroxidase-conjugated secondary antibody for 1 hr and reacted with ECL (Millipore, USA). Finally, the photographs were taken by Tanon-5200 system (Tanon, China). The densitometry of protein bands was quantified by ImageJ 1.51K.
RT-qPCR
Request a detailed protocolThe femurs were collected from wild-type, Mir155-Tg, and Mir155-KO mice. And bones were ground with a high-speed low-temperature tissue homogenizer (Servicebio, China). As previously reported (Teng et al., 2022), the miRNAs were extracted from BMSCs and ground bone tissues with the MolPure Cell/Tissue miRNA Kit (Yeasen, China) as per the manufacturer’s instructions. The miRNA was further reversed by Tailing reaction using miRNA first strand cDNA synthesis kit (Accurate Biology, China). In brief, 3.75 μL miRNA, 5 μL 2×miRNA RT Reaction Solution, 1.25 μL miRNA RT Enzyme Mix were incubated at 37°C for 1 hr, 85°C for 5 min. The mRNAs from osteoclasts induced from Mir155-KO and wild-type mice were extracted with an RNA extraction kit (Accurate Biology, China) as per the manufacturer’s instruction. Total RNA (500 ng) was transcribed with reversed regents (Accurate Biology, China). RT-qPCR was performed using SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology, China) on an AriaMx Real-time quantitative PCR machine (Agilent, USA). The PCR conditions were 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. The fold change relative to the control group was measured by the 2-∆∆Ct method. The primers used were shown in Table 1.
Primers used for RT-qPCR.
| Gene name | Forward sequence (5’->3’) | Reversed sequence (5’->3’) |
|---|---|---|
| Gapdh | TGTGTCCGTCGTGGATCTG | TTGCTGTTGAAGTCGCAGGA |
| Rank | CCAGGAGAGGCATTATGAGCA | ACTGTCGGAGGTAGGAGTGC |
| Ctsk | FCTCGGCGTTTAATTTGGGAGA | TCGAGAGGGAGGTATTCTGAGT |
| Mir155 | 5’-TAATGCTAATTGTGATAGGGGT-3’ | |
PrestoBlue cell viability assay
Request a detailed protocolCell viability was analyzed using PrestoBlue cell viability reagent (Thermo Fisher, USA) (Jaafar et al., 2019). BMSCs (4×103 cells/well) were seeded into 96-well culture plates. After 1, 3, and 5 days, the medium was removed and replaced with a cell viability detection medium according to the manufacturer’s instructions. After 2 hr, the OD value was measured by a microplate reader at 570 nm with a reference wavelength of 600 nm.
TRAP staining
Request a detailed protocolTRAP staining was performed as previously reported (Ho et al., 2017). For osteoclastogenic differentiation, bone marrow cells were cultured in α-minimum essential medium containing 10% fetal bovine serum and added with M-CSF (10 ng/mL) for 4 days. Cells (8×104 cells/well) was supplemented with M-CSF (20 ng/mL) and RANKL (25 ng/mL) for 7 days. TRAP staining was performed with Acid Phosphatase, Leukocyte (TRAP) Kit as the manufacturer’s introduction (Sigma, USA).
ELISA
Request a detailed protocolSerum PINP and CTX-1 are essential markers for evaluating the level of bone resorption (Ye et al., 2021). The levels of PINP and CTX-1 were measured using the PINP ELISA kit (SAB, USA) and CTX-1 ELISA kit (Finebio, China) as the manufacturer’s introduction.
FACS analysis
Request a detailed protocolFACS analysis was performed as previously reported (Zhu et al., 2010). For cell surface staining, Sca-I (1:1000, eBioscience, USA), CD44 (1:1000, eBioscience, USA), CD11b (1:1000, eBioscience, USA), and CD31 (1:200, eBioscience, USA) were used.
Statistical analysis
Request a detailed protocolData are expressed as mean ± SD. Statistical analysis was performed with t-tests for the comparison of the two groups. p<0.05 was considered a significant difference.
Data availability
Source data files have been provided as Figure 1-source data 1, Figure 2-source data 1, Figure 3-source data 1, Figure 4-source data 1, Figure 5-source data 1, Figure 6-source data 1, Figure 7-source data 1, Figure 8-source data 1, Figure 9-source data 1.
References
-
Microrna and cancer -- a brief overviewAdvances in Biological Regulation 57:1–9.https://doi.org/10.1016/j.jbior.2014.09.013
-
Inflammation in bone physiology and pathologyCurrent Opinion in Rheumatology 30:59–64.https://doi.org/10.1097/BOR.0000000000000449
-
Bone health in men with prostate cancer: review articleCurrent Osteoporosis Reports 17:527–537.https://doi.org/10.1007/s11914-019-00536-8
-
Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in miceArthritis and Rheumatism 63:1281–1288.https://doi.org/10.1002/art.30281
-
Inhibition of mir‑155‑5p attenuates the valvular damage induced by rheumatic heart diseaseInternational Journal of Molecular Medicine 45:429–440.https://doi.org/10.3892/ijmm.2019.4420
-
Biologic therapies and systemic bone loss in rheumatoid arthritisAutoimmunity Reviews 12:958–966.https://doi.org/10.1016/j.autrev.2013.03.015
-
The role of micrornas in bone metabolism and diseaseInternational Journal of Molecular Sciences 21:17.https://doi.org/10.3390/ijms21176081
-
The current state of miRNAs as biomarkers and therapeutic toolsClinical and Experimental Medicine 20:349–359.https://doi.org/10.1007/s10238-020-00627-2
-
Mir-155 inhibits mouse osteoblast differentiation by suppressing Smad5 expressionBioMed Research International 2017:1–7.https://doi.org/10.1155/2017/1893520
-
Mir-155 regulates PAD4-dependent formation of neutrophil extracellular trapsFrontiers in Immunology 10:2462.https://doi.org/10.3389/fimmu.2019.02462
-
Physico-mechanical properties of HA/TCP pellets and their three-dimensional biological evaluation in vitroAdvances in Experimental Medicine and Biology 1084:1–15.https://doi.org/10.1007/5584_2017_130
-
Inhibition of miR-155 in MCF-7 breast cancer cell line by gold nanoparticles functionalized with antagomir and AS1411 aptamerJournal of Cellular Physiology 235:6887–6895.https://doi.org/10.1002/jcp.29584
-
Osteocyte-related cytokines regulate osteoclast formation and bone resorptionInternational Journal of Molecular Sciences 21:14.https://doi.org/10.3390/ijms21145169
-
Micrornas as peripheral biomarkers in aging and age-related diseasesProgress in Molecular Biology and Translational Science 146:47–94.https://doi.org/10.1016/bs.pmbts.2016.12.013
-
Improved calvarial bone repair by hascs engineered with cre/loxp-based baculovirus conferring prolonged BMP-2 and mir-148b co-expressionJournal of Tissue Engineering and Regenerative Medicine 11:3068–3077.https://doi.org/10.1002/term.2208
-
MiR-155-5p regulates macrophage M1 polarization and apoptosis in the synovial fluid of patients with knee osteoarthritisExperimental and Therapeutic Medicine 21:68.https://doi.org/10.3892/etm.2020.9500
-
MiR155 modulates vascular calcification by regulating akt-foxo3a signalling and apoptosis in vascular smooth muscle cellsJournal of Cellular and Molecular Medicine 25:535–548.https://doi.org/10.1111/jcmm.16107
-
Microrna-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathwayInternational Journal of Molecular Medicine 41:3379–3393.https://doi.org/10.3892/ijmm.2018.3526
-
Secondary causes of low bone mass in patients with breast cancer: a need for greater vigilanceJournal of Clinical Oncology 27:3605–3610.https://doi.org/10.1200/JCO.2008.20.2549
-
Biology of osteoclast activation in cancerJournal of Clinical Oncology 19:3562–3571.https://doi.org/10.1200/JCO.2001.19.15.3562
-
Sphingosine 1-phosphate receptor activation enhances BMP-2-induced osteoblast differentiationBiochemical and Biophysical Research Communications 423:200–205.https://doi.org/10.1016/j.bbrc.2012.05.130
-
Th17 cell frequency is associated with low bone mass in primary sclerosing cholangitisJournal of Hepatology 70:941–953.https://doi.org/10.1016/j.jhep.2018.12.035
-
Intracellular codelivery of anti-inflammatory drug and anti-MIR 155 to treat inflammatory diseaseActa Pharmaceutica Sinica. B 10:1521–1533.https://doi.org/10.1016/j.apsb.2020.06.005
-
Icariin triggers osteogenic differentiation of bone marrow stem cells by up-regulating mir-335-5pExperimental Cell Research 414:113085.https://doi.org/10.1016/j.yexcr.2022.113085
-
Mir-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cellsCell Death & Differentiation 18:985–995.https://doi.org/10.1038/cdd.2010.167
-
Role of p53/mir-155-5p/sirt1 loop in renal tubular injury of diabetic kidney diseaseJournal of Translational Medicine 16:146.https://doi.org/10.1186/s12967-018-1486-7
-
Roles for mirnas in osteogenic differentiation of bone marrow mesenchymal stem cellsStem Cell Research & Therapy 10:197.https://doi.org/10.1186/s13287-019-1309-7
-
H19X-encoded mir-424 (322) /-503 cluster: emerging roles in cell differentiation, proliferation, plasticity and metabolismCellular and Molecular Life Sciences 76:903–920.https://doi.org/10.1007/s00018-018-2971-0
-
MicroRNA-30c abrogation protects against spinal cord ischemia reperfusion injury through modulating SIRT1European Journal of Pharmacology 851:80–87.https://doi.org/10.1016/j.ejphar.2019.02.027
-
Tshr signaling stimulates proliferation through PI3K/Akt and induction of miR-146a and miR-155 in thyroid eye disease orbital fibroblastsInvestigative Ophthalmology & Visual Science 60:4336–4345.https://doi.org/10.1167/iovs.19-27865
-
Expression of miR-155 and miR-146a in the saliva of patients with periodontitis and its clinical valueAmerican Journal of Translational Research 13:6670–6677.
-
Mir-155 accelerates the growth of human liver cancer cells by activating CDK2 via targeting H3F3AMolecular Therapy Oncolytics 17:471–483.https://doi.org/10.1016/j.omto.2020.05.002
-
Cyclic peptidomimetics as inhibitor for miR-155 biogenesisMolecular Pharmaceutics 16:914–920.https://doi.org/10.1021/acs.molpharmaceut.8b01247
-
Ghrh expression plasmid improves osteoporosis and skin damage in aged miceGrowth Hormone & IGF Research 60–61:101429.https://doi.org/10.1016/j.ghir.2021.101429
-
Sdf-1 mediates mesenchymal stem cell recruitment and migration via the SDF-1/CXCR4 axis in bone defectJournal of Bone and Mineral Metabolism 39:126–138.https://doi.org/10.1007/s00774-020-01122-0
Article and author information
Author details
Funding
The Science and Techonolgoy program of Guangzhou (202201010073)
- Lihong Wu
The Science and Technology program of Guangzhou (202201020116)
- Zhichao Zheng
The National Natural Science Foundation of China (U22A20159)
- Lihong Wu
The National Natural Science Foundation of China (82150410451)
- Janak L Pathak
The General Guiding Project of Guangzhou (20201A011105)
- Zhichao Zheng
The Medical Scientific Research Foundation of Guangdong Province (B2020027)
- Zhichao Zheng
The Undergraduate Science and Technology Innovation Project of Guangzhou Medical University (2020A049)
- Ruoshu Tang
The High-level University Construction Founding of Guangzhou Medical University (06-410-2106035 and 02-412-B205002-1003017)
- Janak L Pathak
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
This project was funded by the Science and Technology program of Guangzhou (202201010073, 202201020116), the National Natural Science Foundation of China (U22A20159, 82150410451), the General Guiding Project of Guangzhou (20201A011105), the Medical Scientific Research Foundation of Guangdong Province (B2020027), the Undergraduate Science and Technology Innovation Project of Guangzhou Medical University (2020A049), and High-level University Construction Funding of Guangzhou Medical University (02-412-B205002-1003017 and 06-410-2106035).
Ethics
The animal experiment was conducted in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China (2017-078).
Copyright
© 2023, Zheng, Wu, Li et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 805
- views
-
- 160
- downloads
-
- 7
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 7
- citations for umbrella DOI https://doi.org/10.7554/eLife.77742






