Host-virus Interactions: Closing the net on retroviruses
HIV-1 like other retroviruses stores its genetic material as RNA, and then converts it to DNA once inside a susceptible host cell. The process of converting RNA to DNA, which is called reverse transcription, takes place inside a protein-based shell called the capsid. The viral DNA is then integrated into the DNA of the host, where it can persist indefinitely.
Host species protect themselves against retroviruses in various ways. TRIM5, for example, is a protein that recognizes the capsid and as a result inhibits reverse transcription (Sayah et al., 2004; Stremlau et al., 2004). Recognition of the capsid lattice by TRIM5 also activates an innate immune response against the virus (Pertel et al., 2011).
TRIM5 belongs to a large family of proteins that have a RING domain, a B-box domain and a coiled-coil domain (Figure 1A). Each of these domains helps the protein to restrict the life cycle of retroviruses (Grütter and Luban, 2012). Moreover, the C-terminal half of TRIM5 binds directly to the capsid (Sebastian and Luban, 2005; Stremlau et al., 2006), and TRIM5 proteins from different species can restrict a range of retroviruses with very different capsids. Now, in two papers in eLife, researchers from the University of Virginia, the University of Utah, Caltech and Ben-Gurion University report new structural insights into how TRIM5 recognizes retroviruses with such diverse capsids (Li et al., 2016; Wagner et al., 2016).
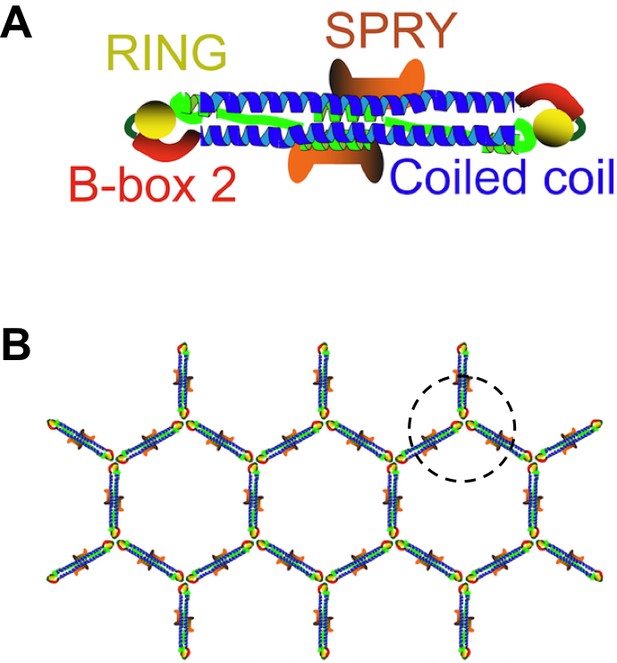
TRIM5 proteins form dimers and a hexagonal lattice.
(A) Most TRIM5 proteins in solution will pair off to form an anti-parallel dimer via their coiled-coil domains (blue). The RING domain (yellow) and B-box domain (red) are at the ends of the dimer, and the C-terminal SPRY domains (orange) are in the center of the dimer. (B) When the SPRY domains bind to the capsid of a retrovirus, the B-box domains of three TRIM5 proteins associate as a trimer (circled). This, in turn, forms a hexagonal lattice of TRIM5 dimers, with the SPRY domains facing into the capsid and the RING domains pointing outwards. Flexibility in the junction between the B-box and coiled-coil domains permits TRIM5 to associate with a wide range of retroviral capsids. Adapted from Figure 1 of Li et al.
TRIM proteins are known to pair together to form dimers via their coiled-coil domains, and these dimers can make a hexagonal lattice that matches the hexagonal lattices found in retroviral capsids (Ganser-Pornillos et al., 2011; Figure 1B). However, these previous studies used artificial TRIM proteins, instead of the naturally occurring TRIM5 protein, because the native protein behaves poorly in vitro. For the same reason, fragments of TRIM proteins were used instead of the full-length version in other experiments to demonstrate that TRIM proteins form anti-parallel dimers rather than parallel ones (Goldstone et al., 2014; Sanchez et al., 2014; Weinert et al., 2015).
In the first paper, Barbie Ganser-Pornillos, Grant Jensen, Wesley Sundquist and colleagues – including Yen-Li Li and Viswanathan Chandrasekaran as joint first authors – report methods that can overcome the problems of working with native TRIM5 protein (Li et al., 2016). Briefly, a flat sheet of capsid lattice (which mimics the capsid of HIV-1) was used to trigger TRIM5 to form its hexagonal lattice under conditions that would otherwise prevent the TRIM5 from doing this spontaneously. Li et al. also showed that this method only worked if the TRIM5 protein could recognize and restrict the viral capsid used in the experiment. The fact that the TRIM5 lattices only form under very specific conditions in vitro suggests that the structures observed are relevant to what happens in vivo.
Li et al. went on to generate stable capsid cores, rather than flat capsid sheets. When they incubated these cores with TRIM5 protein, they saw (via electron cryotomography) that the capsid cores were decorated with hexagonal nets of TRIM5. This demonstrates that native TRIM5 protein forms a hexagonal lattice that matches the lattice of bona fide capsid cores.
In the second paper, Owen Pornillos and colleagues – including Jonathan Wagner as first author – report how the B-box domain of TRIM5 promotes the formation of the hexagonal lattice (Wagner et al., 2016). Crystal structures of the full-length TRIM5 protein have eluded investigators for at least 12 years. However, Wagner et al. generated a B-box domain with a shortened coiled-coil domain and a short linker shaped like a hairpin. They then used this “mini-TRIM” to grow protein crystals, but only after they had confirmed that mini-TRIM behaved like the full-length protein in a number of assays.
The crystal structures showed that the mini-TRIM proteins form both dimers and trimers via their B-box domains (Wagner et al., 2016). This mirrors the observations of another group (Keown et al., 2016). The trimers appear to link TRIM5 into a hexagonal net, which is like the net observed by electron cryotomography (Figure 1B). Wagner et al. showed that the coiled-coil domain can move in relation to the B-box domain; this flexibility could partly explain how a TRIM5 protein from a given species can recognize a diversity of capsid lattice structures.
Many questions remain regarding how the structure of TRIM5 relates to its function. For example, most of the mini-TRIM crystals formed dimers via their B-box domains; do these dimers play a functional role in vivo? Moreover, the antiviral activity of TRIM5 depends on its RING domain and its activity as an E3 ubiquitin ligase (Pertel et al., 2011), but how does recognizing the capsid activate this? Is this activity regulated by the B-box domain and, if so, how? Whatever the case, all the data suggest that the antiviral response of TRIM5 is activated by, and is compatible with, TRIM5 forming trimers and a hexagonal, net-like lattice.
References
-
Hexagonal assembly of a restricting TRIM5α proteinProceedings of the National Academy of Sciences of the United States of America 108:534–539.https://doi.org/10.1073/pnas.1013426108
-
Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid latticeProceedings of the National Academy of Sciences of the United States of America 111:9609–9614.https://doi.org/10.1073/pnas.1402448111
-
TRIM5 structure, HIV-1 capsid recognition, and innate immune signalingCurrent Opinion in Virology 2:142–150.https://doi.org/10.1016/j.coviro.2012.02.003
-
The tripartite motif coiled-coil is an elongated antiparallel hairpin dimerProceedings of the National Academy of Sciences of the United States of America 111:2494–2499.https://doi.org/10.1073/pnas.1318962111
-
Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factorProceedings of the National Academy of Sciences of the United States of America 103:5514–5519.https://doi.org/10.1073/pnas.0509996103
Article and author information
Author details
Publication history
Copyright
© 2016, Luban
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,168
- views
-
- 166
- downloads
-
- 1
- citation
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 1
- citation for umbrella DOI https://doi.org/10.7554/eLife.18243
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Biochemistry and Chemical Biology
- Structural Biology and Molecular Biophysics
Restriction factors and pattern recognition receptors are important components of intrinsic cellular defenses against viral infection. Mammalian TRIM5α proteins are restriction factors and receptors that target the capsid cores of retroviruses and activate ubiquitin-dependent antiviral responses upon capsid recognition. Here, we report crystallographic and functional studies of the TRIM5α B-box 2 domain, which mediates higher-order assembly of TRIM5 proteins. The B-box can form both dimers and trimers, and the trimers can link multiple TRIM5α proteins into a hexagonal net that matches the lattice arrangement of capsid subunits and enables avid capsid binding. Two modes of conformational flexibility allow TRIM5α to accommodate the variable curvature of retroviral capsids. B-box mediated interactions also modulate TRIM5α’s E3 ubiquitin ligase activity, by stereochemically restricting how the N-terminal RING domain can dimerize. Overall, these studies define important molecular details of cellular recognition of retroviruses, and how recognition links to downstream processes to disable the virus.






