Highlights
-

Equity, diversion, inclusion
eLife has published its seventh report on its work to address inequities in research and publishing.
-

1000 Insights and counting
From mapping the brain of fruit flies to the science behind kombucha, we look back at more than ten years of Insight articles.
-
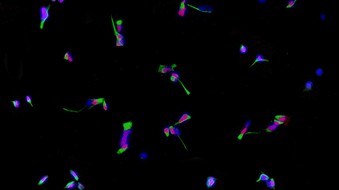
mTOR signaling and autism
Genetically distinct subtypes of autism have common neurite-related defects that are driven by alterations in mTOR signaling.
Magazine
Research categories
- Biochemistry and Chemical Biology
- Cancer Biology
- Cell Biology
- Chromosomes and Gene Expression
- Computational and Systems Biology
- Developmental Biology
- Ecology
- Epidemiology and Global Health
- Evolutionary Biology
- Genetics and Genomics
- Immunology and Inflammation
- Medicine
- Microbiology and Infectious Disease
- Neuroscience
- Physics of Living Systems
- Plant Biology
- Stem Cells and Regenerative Medicine
- Structural Biology and Molecular Biophysics





